hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
d0b3fc66ce0d6bd9032a470f59ae7cc79efcef37 | 380,275 | ipynb | Jupyter Notebook | notebooks/qos_analysis.ipynb | Abellan09/tfg_crawler_i2p | 83e4e0a5548727dbf184ef34b96ca0f5b7636dc8 | [
"MIT"
] | 4 | 2018-06-19T20:18:10.000Z | 2018-09-02T15:58:21.000Z | notebooks/qos_analysis.ipynb | Abellan09/tfg_crawler_i2p | 83e4e0a5548727dbf184ef34b96ca0f5b7636dc8 | [
"MIT"
] | null | null | null | notebooks/qos_analysis.ipynb | Abellan09/tfg_crawler_i2p | 83e4e0a5548727dbf184ef34b96ca0f5b7636dc8 | [
"MIT"
] | 3 | 2019-01-18T16:33:34.000Z | 2020-01-28T02:13:27.000Z | 233.154506 | 197,908 | 0.880978 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"# Reading QoS analysis raw info\nTemporarily, this info is saved in a CSV file but it will be in the database\n\n**qos_analysis_13112018.csv**\n- columns = ['url','protocol','code','start','end','duration','runid']\n- First try of qos analysis.\n- It was obtained from 50 repetitions of each of 3921 eepsites gathered. \n- Just one i2router (UCA desktop host)\n- Time gap between each eepsite request 3921*0.3sec=1176sec/60sec ~ 19 mins\n- Total experiment elapsed time 50rep X 19 mins ~ 16 hours\n\n**qos_analysis_29112018_local.csv**\n\n- columns = ['url','code','duration','runid']\n- 100 repetions of the first 10 eepsite from the list. Just for testing.\n- local i2prouter from my laptop\n- Time gap between each eepsite 10*5sec=50sec ~ 60s\n- Total experiment elapsed time 100rep x 1min ~ 100 mins\n\n",
"_____no_output_____"
]
],
[
[
"# File for processing it\nqos_file = 'qos_analysis_13112018.csv'\npath_to_file = 'data/' + qos_file\n\ncolumns = ['url','protocol','code','start','end','duration','runid']\ndf_qos = pd.read_csv(path_to_file,names=columns,delimiter=\"|\")\n\n# File for processing it - local router\nqos_file = 'qos_analysis_29112018_local.csv'\npath_to_file = 'data/' + qos_file\ncolumns = ['url','code','duration','runid']\ndf_qos_local = pd.read_csv(path_to_file,names=columns,delimiter=\"|\")\n\n# File for processing it - local router\nqos_file = 'qos_analysis_29112018_remote.csv'\npath_to_file = 'data/' + qos_file\ncolumns = ['url','code','duration','runid']\ndf_qos_remote = pd.read_csv(path_to_file,names=columns,delimiter=\"|\")\n\n# File for testing - to be removed\nqos_file = 'analitica.csv'\npath_to_file = 'data/' + qos_file\ncolumns = ['url','code','duration','runid','intervals']\ndf_qos_testing = pd.read_csv(path_to_file,names=columns,delimiter=\"|\")",
"_____no_output_____"
],
[
"# DF to analize\ndf_qos = df_qos_testing.copy()\n\n# Removing not valid rounds\ndf_qos['runid'] = pd.to_numeric(df_qos['runid'], errors='coerce').dropna()\n",
"_____no_output_____"
],
[
"df_qos.head()",
"_____no_output_____"
],
[
"# Duration distribution by http response\nfig, ax1 = plt.subplots(figsize=(10, 6))\n\n# http code\ncode = 200\n\ndf_to_plot = df_qos[(df_qos['code']==code)]['duration']\n#df_qos[(df_qos['code']==500)]['duration'].hist(bins=100)\n\ndf_to_plot.plot(kind='hist',bins=100, ax=ax1, color={'r','g'}, alpha=0.7)\nax1.set_ylabel('Frequency')\nax1.set_xlabel('Duration (seconds)')\nax1.set_title('HTTP ' + str(code))\nplt.sca(ax1)# matplotlib only acts over the current axis\nplt.xticks(rotation=75)",
"_____no_output_____"
],
[
"df_qos['code'].hist(bins=100)",
"_____no_output_____"
],
[
"df_qos['code'].unique()",
"_____no_output_____"
],
[
"df_qos.code.value_counts()",
"_____no_output_____"
],
[
"df_qos.describe()",
"_____no_output_____"
],
[
"# Average duration by error code\ndf = pd.DataFrame({\n 'code': df_qos['code'],\n 'duration': df_qos['duration'],\n})\n\ndf = df.sort_values(by='code')\n\nfig, ax1 = plt.subplots(figsize=(12, 8))\n\nto_drop = []\n\ndf = df[~df['code'].isin(to_drop)]\n\nmeans = df.groupby('code').mean()\nstd = df.groupby('code').std()\n\nmeans.plot(kind='bar',yerr=std, ax=ax1, color={'r','g'}, alpha=0.7)\nax1.set_ylabel('Duration average (seconds)')\nplt.sca(ax1)# matplotlib only acts over the current axis\nplt.xticks(rotation=75)\n",
"_____no_output_____"
],
[
"df.groupby('code').describe()",
"_____no_output_____"
],
[
"# Duration by error code\ndf = pd.DataFrame({\n 'code': df_qos['code'],\n 'duration': df_qos['duration'],\n})\n\nto_drop = [504]\n\ndf = df[~df['code'].isin(to_drop)]\n\nfig, ax1 = plt.subplots(figsize=(12, 8))\n\nax = sns.boxplot(x=\"code\", y=\"duration\", data=df, ax=ax1)\nax1.set_ylabel('Duration (seconds)')\nax1.set_xticklabels(set(df.code))\nplt.sca(ax1)# matplotlib",
"_____no_output_____"
],
[
"# Average duration by eepsite\ndf = pd.DataFrame({\n 'url': df_qos['url'],\n 'duration': df_qos['duration'],\n})\n\nfig, ax1 = plt.subplots(figsize=(15, 8),)\n\ndf = df.sort_values(by='url')\n\nmeans = df.groupby('url').mean()\nstd = df.groupby('url').std()\n\nmeans = means[0:50]\nstd = std[0:50]\n\nmeans.plot(kind='bar',yerr=std, ax=ax1, color={'r'}, alpha=0.7)\nax1.set_ylabel('Duration average (seconds)')\nplt.sca(ax1)# matplotlib only acts over the current axis\nplt.xticks(rotation=90)",
"_____no_output_____"
],
[
"# Average duration by eepsite\ndf = pd.DataFrame({\n 'url': df_qos['url'],\n 'duration': df_qos['duration'],\n 'code': df_qos['code']\n})\n\nfig, ax1 = plt.subplots(figsize=(15, 8),)\n\ndf = df.sort_values(by='duration',ascending=False)\n\neepsites = list(df[0:10000].groupby('url').groups.keys())[0:20]\ndf = df[df['url'].isin(eepsites)]\n\nax = sns.boxplot(x=\"url\", y=\"duration\", data=df, hue='code', ax=ax1)\nax1.set_ylabel('Duration (seconds)')\n#ax1.set_ylim((0,3))\nplt.sca(ax1)# matplotlib only acts over the current axis\nplt.xticks(rotation=90)",
"_____no_output_____"
]
],
[
[
"# Availability study",
"_____no_output_____"
]
],
[
[
"HTTP_RESPONSE_CODES = {200:'OK', \n 301:'Moved Permanently', \n 302:'Found (Previously \"Moved temporarily\")', \n 400:'Bad Request', \n 401:'Unauthorized',\n 403:'Forbidden',\n 429:'Too Many Requests',\n 500:'Internal Server Error',\n 502:'Bad Gateway',\n 503:'Service Unavailable',\n 504:'Gateway Timeout'}",
"_____no_output_____"
],
[
"df_qos",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0b4019a51fef66f367402c360751fdfe4f00d6d | 187,406 | ipynb | Jupyter Notebook | user_complaints/NLP_complaints.ipynb | ffzmm/user_complaints | 5840020fc087f315cc951e9d74ba9d7630b0475f | [
"Apache-2.0"
] | null | null | null | user_complaints/NLP_complaints.ipynb | ffzmm/user_complaints | 5840020fc087f315cc951e9d74ba9d7630b0475f | [
"Apache-2.0"
] | null | null | null | user_complaints/NLP_complaints.ipynb | ffzmm/user_complaints | 5840020fc087f315cc951e9d74ba9d7630b0475f | [
"Apache-2.0"
] | null | null | null | 280.968516 | 160,509 | 0.667257 | [
[
[
"import pandas as pd\ndf = pd.read_csv('data/Consumer_Complaints.csv')\ndf.info()",
"/anaconda3/lib/python3.6/site-packages/IPython/core/interactiveshell.py:2728: DtypeWarning: Columns (5,6,11,16) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"feature_col = ['Consumer complaint narrative']\nres_col = ['Product', 'Issue']",
"_____no_output_____"
],
[
"df.dropna(subset= feature_col + res_col, inplace=True)\ndf.drop_duplicates(subset=feature_col, inplace=True)\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 367629 entries, 44142 to 929033\nData columns (total 18 columns):\nDate received 367629 non-null object\nProduct 367629 non-null object\nSub-product 317356 non-null object\nIssue 367629 non-null object\nSub-issue 254272 non-null object\nConsumer complaint narrative 367629 non-null object\nCompany public response 173475 non-null object\nCompany 367629 non-null object\nState 366264 non-null object\nZIP code 281945 non-null object\nTags 64498 non-null object\nConsumer consent provided? 367629 non-null object\nSubmitted via 367629 non-null object\nDate sent to company 367629 non-null object\nCompany response to consumer 367625 non-null object\nTimely response? 367629 non-null object\nConsumer disputed? 160949 non-null object\nComplaint ID 367629 non-null int64\ndtypes: int64(1), object(17)\nmemory usage: 53.3+ MB\n"
],
[
"#print(df['Product'].unique())",
"_____no_output_____"
],
[
"df_cat = None\nfor col in res_col:\n temp = df[col].astype('category')\n df_cat = pd.concat([df_cat, temp], axis=1)\n\ndf.drop(columns=res_col, inplace=True)\ndf = pd.concat([df, df_cat], axis=1)\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 367629 entries, 44142 to 929033\nData columns (total 18 columns):\nDate received 367629 non-null object\nSub-product 317356 non-null object\nSub-issue 254272 non-null object\nConsumer complaint narrative 367629 non-null object\nCompany public response 173475 non-null object\nCompany 367629 non-null object\nState 366264 non-null object\nZIP code 281945 non-null object\nTags 64498 non-null object\nConsumer consent provided? 367629 non-null object\nSubmitted via 367629 non-null object\nDate sent to company 367629 non-null object\nCompany response to consumer 367625 non-null object\nTimely response? 367629 non-null object\nConsumer disputed? 160949 non-null object\nComplaint ID 367629 non-null int64\nProduct 367629 non-null category\nIssue 367629 non-null category\ndtypes: category(2), int64(1), object(15)\nmemory usage: 48.7+ MB\n"
],
[
"# print(df['Issue'].unique())",
"_____no_output_____"
],
[
"# randomly select train/test data\nfrom sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(df[feature_col[0]], df[res_col[0]], test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"y_train.head()\n#X_train.head()",
"_____no_output_____"
],
[
"import nltk\nnltk.download('wordnet')\nfrom nltk.stem.wordnet import WordNetLemmatizer\nimport gensim\nfrom gensim.utils import simple_preprocess\nimport gensim.corpora as corpora",
"[nltk_data] Downloading package wordnet to /Users/yunfei/nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n"
],
[
"from nltk.corpus import stopwords\nstop_words = stopwords.words('english')\nstop_words.extend(['xxxx', 'xx'])\nstop_words = set(stop_words)",
"_____no_output_____"
],
[
"def tokenize(doc):\n # doc is a string\n # return an array of words\n return simple_preprocess(doc, deacc=True, min_len=2, max_len=15)\n\ndef rm_stopwords_and_lemmatize(token_array, flag_rm_stop=True, flag_lemmatize=True):\n out = []\n for token in token_array:\n if flag_lemmatize:\n token = WordNetLemmatizer().lemmatize(token)\n if flag_rm_stop:\n if token not in stop_words:\n out.append(token)\n else:\n out.append(token)\n return out\n\ndef my_tokenizer(doc, flag_rm_stop=True, flag_lemmatize=True):\n return rm_stopwords_and_lemmatize(tokenize(doc), flag_rm_stop, flag_lemmatize)",
"_____no_output_____"
],
[
"#text = 'I struggled so much with the settings.'\n#tokens = tokenize(text)\n#print(tokens)\n#print(rm_stopwords_and_lemmatize(tokens))\n#print(rm_stopwords_and_lemmatize(tokens, flag_rm_stop=False))\n#print(rm_stopwords_and_lemmatize(tokens, flag_lemmatize=False))\n#print(rm_stopwords_and_lemmatize(tokens, flag_rm_stop=False, flag_lemmatize=False))\n#print(my_tokenizer(text))",
"_____no_output_____"
],
[
"#define vectorizer parameters\nfrom sklearn.feature_extraction.text import TfidfVectorizer\n\ntfidf_vectorizer = TfidfVectorizer(max_features=1000,\n min_df=5, \n stop_words='english',\n use_idf=True, \n tokenizer=my_tokenizer, \n token_pattern=r\"\\b\\w[\\w']+\\b\",\n ngram_range=(1,2))\n\ntfidf_matrix = tfidf_vectorizer.fit_transform(X_train) #fit the vectorizer to corpus (min = 0.0, max = 1.0)\n\nprint (tfidf_matrix.shape)",
"(294103, 1000)\n"
],
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\n\nclf = LogisticRegression(random_state=0, solver='lbfgs', multi_class='multinomial')\n#clf = RandomForestClassifier()",
"_____no_output_____"
],
[
"from sklearn.model_selection import cross_val_score\n\nacc = cross_val_score(clf, tfidf_matrix, y_train, scoring='accuracy', cv=5)\nprint(acc)",
"[0.6764066 0.67637909 0.67902683 0.67967355 0.67703817]\n"
],
[
"#model = clf.fit(tfidf_matrix, y_train)",
"_____no_output_____"
],
[
"from sklearn.decomposition import LatentDirichletAllocation\n\nn_topics = 20\nlda = LatentDirichletAllocation(n_components=n_topics, \n learning_method='online')",
"_____no_output_____"
],
[
"tfidf_matrix_lda = (tfidf_matrix * 100)\ntfidf_matrix_lda = tfidf_matrix_lda.astype(int)",
"_____no_output_____"
],
[
"lda.fit(tfidf_matrix_lda)",
"_____no_output_____"
],
[
"print(lda.components_.shape)",
"(20, 1000)\n"
],
[
"import pyLDAvis\nimport pyLDAvis.sklearn\npyLDAvis.enable_notebook()\n\npyLDAvis.sklearn.prepare(lda, tfidf_matrix_lda, tfidf_vectorizer)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b402b3c69a8034724919b1988ce26f2e9f688c | 33,043 | ipynb | Jupyter Notebook | advanced_functionality/distributed_tensorflow_mask_rcnn/mask-rcnn-scriptmode-efs.ipynb | fhirschmann/amazon-sagemaker-examples | bb4a4ed78cd4f3673bd6894f0b92ab08aa7f8f29 | [
"Apache-2.0"
] | null | null | null | advanced_functionality/distributed_tensorflow_mask_rcnn/mask-rcnn-scriptmode-efs.ipynb | fhirschmann/amazon-sagemaker-examples | bb4a4ed78cd4f3673bd6894f0b92ab08aa7f8f29 | [
"Apache-2.0"
] | null | null | null | advanced_functionality/distributed_tensorflow_mask_rcnn/mask-rcnn-scriptmode-efs.ipynb | fhirschmann/amazon-sagemaker-examples | bb4a4ed78cd4f3673bd6894f0b92ab08aa7f8f29 | [
"Apache-2.0"
] | null | null | null | 40.394866 | 529 | 0.540114 | [
[
[
"# Distributed Training of Mask-RCNN in Amazon SageMaker using EFS\n\nThis notebook is a step-by-step tutorial on distributed training of [Mask R-CNN](https://arxiv.org/abs/1703.06870) implemented in [TensorFlow](https://www.tensorflow.org/) framework. Mask R-CNN is also referred to as heavy weight object detection model and it is part of [MLPerf](https://www.mlperf.org/training-results-0-6/).\n\nConcretely, we will describe the steps for training [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) and [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) in [Amazon SageMaker](https://aws.amazon.com/sagemaker/) using [Amazon EFS](https://aws.amazon.com/efs/) file-system as data source.\n\nThe outline of steps is as follows:\n\n1. Stage COCO 2017 dataset in [Amazon S3](https://aws.amazon.com/s3/)\n2. Copy COCO 2017 dataset from S3 to Amazon EFS file-system mounted on this notebook instance\n3. Build Docker training image and push it to [Amazon ECR](https://aws.amazon.com/ecr/)\n4. Configure data input channels\n5. Configure hyper-prarameters\n6. Define training metrics\n7. Define training job and start training\n\nBefore we get started, let us initialize two python variables ```aws_region``` and ```s3_bucket``` that we will use throughout the notebook. The ```s3_bucket``` must be located in the region of this notebook instance.",
"_____no_output_____"
]
],
[
[
"import boto3\n\nsession = boto3.session.Session()\naws_region = session.region_name\ns3_bucket = # your-s3-bucket-name\n\n\ntry:\n s3_client = boto3.client('s3')\n response = s3_client.get_bucket_location(Bucket=s3_bucket)\n print(f\"Bucket region: {response['LocationConstraint']}\")\nexcept:\n print(f\"Access Error: Check if '{s3_bucket}' S3 bucket is in '{aws_region}' region\")",
"_____no_output_____"
]
],
[
[
"## Stage COCO 2017 dataset in Amazon S3\n\nWe use [COCO 2017 dataset](http://cocodataset.org/#home) for training. We download COCO 2017 training and validation dataset to this notebook instance, extract the files from the dataset archives, and upload the extracted files to your Amazon [S3 bucket](https://docs.aws.amazon.com/AmazonS3/latest/gsg/CreatingABucket.html). The ```prepare-s3-bucket.sh``` script executes this step. ",
"_____no_output_____"
]
],
[
[
"!cat ./prepare-s3-bucket.sh",
"_____no_output_____"
]
],
[
[
"Using your *Amazon S3 bucket* as argument, run the cell below. If you have already uploaded COCO 2017 dataset to your Amazon S3 bucket, you may skip this step. ",
"_____no_output_____"
]
],
[
[
"%%time\n!./prepare-s3-bucket.sh {s3_bucket}",
"_____no_output_____"
]
],
[
[
"## Copy COCO 2017 dataset from S3 to Amazon EFS\n\nNext, we copy COCO 2017 dataset from S3 to EFS file-system. The ```prepare-efs.sh``` script executes this step.",
"_____no_output_____"
]
],
[
[
"!cat ./prepare-efs.sh",
"_____no_output_____"
]
],
[
[
"If you have already copied COCO 2017 dataset from S3 to your EFS file-system, skip this step.",
"_____no_output_____"
]
],
[
[
"%%time\n!./prepare-efs.sh {s3_bucket}",
"_____no_output_____"
]
],
[
[
"## Build and push SageMaker training images\n\nFor this step, the [IAM Role](https://docs.aws.amazon.com/IAM/latest/UserGuide/id_roles.html) attached to this notebook instance needs full access to Amazon ECR service. If you created this notebook instance using the ```./stack-sm.sh``` script in this repository, the IAM Role attached to this notebook instance is already setup with full access to ECR service. \n\nBelow, we have a choice of two different implementations:\n\n1. [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) implementation supports a maximum per-GPU batch size of 1, and does not support mixed precision. It can be used with mainstream TensorFlow releases.\n\n2. [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) is an optimized implementation that supports a maximum batch size of 4 and supports mixed precision. This implementation uses custom TensorFlow ops. The required custom TensorFlow ops are available in [AWS Deep Learning Container](https://github.com/aws/deep-learning-containers/blob/master/available_images.md) images in ```tensorflow-training``` repository with image tag ```1.15.2-gpu-py36-cu100-ubuntu18.04```, or later.\n\nIt is recommended that you build and push both SageMaker training images and use either image for training later.",
"_____no_output_____"
],
[
"### TensorPack Faster-RCNN/Mask-RCNN\n\nUse ```./container-script-mode/build_tools/build_and_push.sh``` script to build and push the TensorPack Faster-RCNN/Mask-RCNN training image to Amazon ECR.",
"_____no_output_____"
]
],
[
[
"!cat ./container-script-mode/build_tools/build_and_push.sh",
"_____no_output_____"
],
[
"%%time\n! ./container-script-mode/build_tools/build_and_push.sh {aws_region}",
"_____no_output_____"
]
],
[
[
"Set ```tensorpack_image``` below to Amazon ECR URI of the image you pushed above.",
"_____no_output_____"
]
],
[
[
"tensorpack_image = #<amazon-ecr-uri>",
"_____no_output_____"
]
],
[
[
"### AWS Samples Mask R-CNN\nUse ```./container-optimized-script-mode/build_tools/build_and_push.sh``` script to build and push the AWS Samples Mask R-CNN training image to Amazon ECR.",
"_____no_output_____"
]
],
[
[
"!cat ./container-optimized-script-mode/build_tools/build_and_push.sh",
"_____no_output_____"
]
],
[
[
"Using your *AWS region* as argument, run the cell below. ",
"_____no_output_____"
]
],
[
[
"%%time\n! ./container-optimized-script-mode/build_tools/build_and_push.sh {aws_region}",
"_____no_output_____"
]
],
[
[
"Set ```aws_samples_image``` below to Amazon ECR URI of the image you pushed above.",
"_____no_output_____"
]
],
[
[
"aws_samples_image = #<amazon-ecr-uri> ",
"_____no_output_____"
]
],
[
[
"### Upgrade SageMaker Python SDK\n\nIf needed, upgrade SageMaker Python SDK.",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade pip\n!pip install sagemaker",
"_____no_output_____"
]
],
[
[
"## SageMaker Initialization \n\nWe have staged the data and we have built and pushed the training docker image to Amazon ECR. Now we are ready to start using Amazon SageMaker. ",
"_____no_output_____"
]
],
[
[
"%%time\nimport os\nimport time\nimport sagemaker\nfrom sagemaker import get_execution_role\nfrom sagemaker.tensorflow.estimator import TensorFlow\n\nrole = get_execution_role() # provide a pre-existing role ARN as an alternative to creating a new role\nprint(f'SageMaker Execution Role:{role}')\n\nclient = boto3.client('sts')\naccount = client.get_caller_identity()['Account']\nprint(f'AWS account:{account}')",
"_____no_output_____"
]
],
[
[
"Next, we set the Amazon ECR image URI used for training. You saved this URI in a previous step.",
"_____no_output_____"
]
],
[
[
"training_image = # set to tensorpack_image or aws_samples_image \nprint(f'Training image: {training_image}')",
"_____no_output_____"
]
],
[
[
"## Define SageMaker Data Channels\n\nNext, we define the *train* and *log* data channels using EFS file-system. To do so, we need to specify the EFS file-system id, which is shown in the output of the command below.",
"_____no_output_____"
]
],
[
[
"notebook_attached_efs=!df -kh | grep 'fs-' | sed 's/\\(fs-[0-9a-z]*\\).*/\\1/'\nprint(f\"SageMaker notebook attached EFS: {notebook_attached_efs}\")",
"_____no_output_____"
]
],
[
[
"In the cell below, we define the `train` data input channel.",
"_____no_output_____"
]
],
[
[
"from sagemaker.inputs import FileSystemInput\n\n# Specify EFS file system id.\nfile_system_id = notebook_attached_efs[0]\nprint(f\"EFS file-system-id: {file_system_id}\")\n\n# Specify directory path for input data on the file system. \n# You need to provide normalized and absolute path below.\nfile_system_directory_path = '/mask-rcnn/sagemaker/input/train'\nprint(f'EFS file-system data input path: {file_system_directory_path}')\n\n# Specify the access mode of the mount of the directory associated with the file system. \n# Directory must be mounted 'ro'(read-only).\nfile_system_access_mode = 'ro'\n\n# Specify your file system type\nfile_system_type = 'EFS'\n\ntrain = FileSystemInput(file_system_id=file_system_id,\n file_system_type=file_system_type,\n directory_path=file_system_directory_path,\n file_system_access_mode=file_system_access_mode)\n",
"_____no_output_____"
]
],
[
[
"Below we create the log output directory and define the `log` data output channel.",
"_____no_output_____"
]
],
[
[
"# Specify directory path for log output on the EFS file system.\n# You need to provide normalized and absolute path below.\n# For example, '/mask-rcnn/sagemaker/output/log'\n# Log output directory must not exist\nfile_system_directory_path = f'/mask-rcnn/sagemaker/output/log-{int(time.time())}'\n\n# Create the log output directory. \n# EFS file-system is mounted on '$HOME/efs' mount point for this notebook.\nhome_dir=os.environ['HOME']\nlocal_efs_path = os.path.join(home_dir,'efs', file_system_directory_path[1:])\nprint(f\"Creating log directory on EFS: {local_efs_path}\")\n\nassert not os.path.isdir(local_efs_path)\n! sudo mkdir -p -m a=rw {local_efs_path}\nassert os.path.isdir(local_efs_path)\n\n# Specify the access mode of the mount of the directory associated with the file system. \n# Directory must be mounted 'rw'(read-write).\nfile_system_access_mode = 'rw'\n\n\nlog = FileSystemInput(file_system_id=file_system_id,\n file_system_type=file_system_type,\n directory_path=file_system_directory_path,\n file_system_access_mode=file_system_access_mode)\n\ndata_channels = {'train': train, 'log': log}",
"_____no_output_____"
]
],
[
[
"Next, we define the model output location in S3. Set ```s3_bucket``` to your S3 bucket name prior to running the cell below. \n\nThe model checkpoints, logs and Tensorboard events will be written to the log output directory on the EFS file system you created above. At the end of the model training, they will be copied from the log output directory to the `s3_output_location` defined below.",
"_____no_output_____"
]
],
[
[
"prefix = \"mask-rcnn/sagemaker\" #prefix in your bucket\ns3_output_location = f's3://{s3_bucket}/{prefix}/output'\nprint(f'S3 model output location: {s3_output_location}')",
"_____no_output_____"
]
],
[
[
"## Configure Hyper-parameters\n\nNext we define the hyper-parameters. \n\nNote, some hyper-parameters are different between the two implementations. The batch size per GPU in TensorPack Faster-RCNN/Mask-RCNN is fixed at 1, but is configurable in AWS Samples Mask-RCNN. The learning rate schedule is specified in units of steps in TensorPack Faster-RCNN/Mask-RCNN, but in epochs in AWS Samples Mask-RCNN.\n\nThe detault learning rate schedule values shown below correspond to training for a total of 24 epochs, at 120,000 images per epoch.\n\n<table align='left'>\n <caption>TensorPack Faster-RCNN/Mask-RCNN Hyper-parameters</caption>\n <tr>\n <th style=\"text-align:center\">Hyper-parameter</th>\n <th style=\"text-align:center\">Description</th>\n <th style=\"text-align:center\">Default</th>\n </tr>\n <tr>\n <td style=\"text-align:center\">mode_fpn</td>\n <td style=\"text-align:left\">Flag to indicate use of Feature Pyramid Network (FPN) in the Mask R-CNN model backbone</td>\n <td style=\"text-align:center\">\"True\"</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">mode_mask</td>\n <td style=\"text-align:left\">A value of \"False\" means Faster-RCNN model, \"True\" means Mask R-CNN moodel</td>\n <td style=\"text-align:center\">\"True\"</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">eval_period</td>\n <td style=\"text-align:left\">Number of epochs period for evaluation during training</td>\n <td style=\"text-align:center\">1</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">lr_schedule</td>\n <td style=\"text-align:left\">Learning rate schedule in training steps</td>\n <td style=\"text-align:center\">'[240000, 320000, 360000]'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">batch_norm</td>\n <td style=\"text-align:left\">Batch normalization option ('FreezeBN', 'SyncBN', 'GN', 'None') </td>\n <td style=\"text-align:center\">'FreezeBN'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">images_per_epoch</td>\n <td style=\"text-align:left\">Images per epoch </td>\n <td style=\"text-align:center\">120000</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">data_train</td>\n <td style=\"text-align:left\">Training data under data directory</td>\n <td style=\"text-align:center\">'coco_train2017'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">data_val</td>\n <td style=\"text-align:left\">Validation data under data directory</td>\n <td style=\"text-align:center\">'coco_val2017'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">resnet_arch</td>\n <td style=\"text-align:left\">Must be 'resnet50' or 'resnet101'</td>\n <td style=\"text-align:center\">'resnet50'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">backbone_weights</td>\n <td style=\"text-align:left\">ResNet backbone weights</td>\n <td style=\"text-align:center\">'ImageNet-R50-AlignPadding.npz'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">load_model</td>\n <td style=\"text-align:left\">Pre-trained model to load</td>\n <td style=\"text-align:center\"></td>\n </tr>\n <tr>\n <td style=\"text-align:center\">config:</td>\n <td style=\"text-align:left\">Any hyperparamter prefixed with <b>config:</b> is set as a model config parameter</td>\n <td style=\"text-align:center\"></td>\n </tr>\n</table>\n\n \n<table align='left'>\n <caption>AWS Samples Mask-RCNN Hyper-parameters</caption>\n <tr>\n <th style=\"text-align:center\">Hyper-parameter</th>\n <th style=\"text-align:center\">Description</th>\n <th style=\"text-align:center\">Default</th>\n </tr>\n <tr>\n <td style=\"text-align:center\">mode_fpn</td>\n <td style=\"text-align:left\">Flag to indicate use of Feature Pyramid Network (FPN) in the Mask R-CNN model backbone</td>\n <td style=\"text-align:center\">\"True\"</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">mode_mask</td>\n <td style=\"text-align:left\">A value of \"False\" means Faster-RCNN model, \"True\" means Mask R-CNN moodel</td>\n <td style=\"text-align:center\">\"True\"</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">eval_period</td>\n <td style=\"text-align:left\">Number of epochs period for evaluation during training</td>\n <td style=\"text-align:center\">1</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">lr_epoch_schedule</td>\n <td style=\"text-align:left\">Learning rate schedule in epochs</td>\n <td style=\"text-align:center\">'[(16, 0.1), (20, 0.01), (24, None)]'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">batch_size_per_gpu</td>\n <td style=\"text-align:left\">Batch size per gpu ( Minimum 1, Maximum 4)</td>\n <td style=\"text-align:center\">4</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">batch_norm</td>\n <td style=\"text-align:left\">Batch normalization option ('FreezeBN', 'SyncBN', 'GN', 'None') </td>\n <td style=\"text-align:center\">'FreezeBN'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">images_per_epoch</td>\n <td style=\"text-align:left\">Images per epoch </td>\n <td style=\"text-align:center\">120000</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">data_train</td>\n <td style=\"text-align:left\">Training data under data directory</td>\n <td style=\"text-align:center\">'train2017'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">backbone_weights</td>\n <td style=\"text-align:left\">ResNet backbone weights</td>\n <td style=\"text-align:center\">'ImageNet-R50-AlignPadding.npz'</td>\n </tr>\n <tr>\n <td style=\"text-align:center\">load_model</td>\n <td style=\"text-align:left\">Pre-trained model to load</td>\n <td style=\"text-align:center\"></td>\n </tr>\n <tr>\n <td style=\"text-align:center\">config:</td>\n <td style=\"text-align:left\">Any hyperparamter prefixed with <b>config:</b> is set as a model config parameter</td>\n <td style=\"text-align:center\"></td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"hyperparameters = {\n \"mode_fpn\": \"True\",\n \"mode_mask\": \"True\",\n \"eval_period\": 1,\n \"batch_norm\": \"FreezeBN\"\n }",
"_____no_output_____"
]
],
[
[
"## Define Training Metrics\nNext, we define the regular expressions that SageMaker uses to extract algorithm metrics from training logs and send them to [AWS CloudWatch metrics](https://docs.aws.amazon.com/en_pv/AmazonCloudWatch/latest/monitoring/working_with_metrics.html). These algorithm metrics are visualized in SageMaker console.",
"_____no_output_____"
]
],
[
[
"metric_definitions=[\n {\n \"Name\": \"fastrcnn_losses/box_loss\",\n \"Regex\": \".*fastrcnn_losses/box_loss:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"fastrcnn_losses/label_loss\",\n \"Regex\": \".*fastrcnn_losses/label_loss:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"fastrcnn_losses/label_metrics/accuracy\",\n \"Regex\": \".*fastrcnn_losses/label_metrics/accuracy:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"fastrcnn_losses/label_metrics/false_negative\",\n \"Regex\": \".*fastrcnn_losses/label_metrics/false_negative:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"fastrcnn_losses/label_metrics/fg_accuracy\",\n \"Regex\": \".*fastrcnn_losses/label_metrics/fg_accuracy:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"fastrcnn_losses/num_fg_label\",\n \"Regex\": \".*fastrcnn_losses/num_fg_label:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"maskrcnn_loss/accuracy\",\n \"Regex\": \".*maskrcnn_loss/accuracy:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"maskrcnn_loss/fg_pixel_ratio\",\n \"Regex\": \".*maskrcnn_loss/fg_pixel_ratio:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"maskrcnn_loss/maskrcnn_loss\",\n \"Regex\": \".*maskrcnn_loss/maskrcnn_loss:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"maskrcnn_loss/pos_accuracy\",\n \"Regex\": \".*maskrcnn_loss/pos_accuracy:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(bbox)/IoU=0.5\",\n \"Regex\": \".*mAP\\\\(bbox\\\\)/IoU=0\\\\.5:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(bbox)/IoU=0.5:0.95\",\n \"Regex\": \".*mAP\\\\(bbox\\\\)/IoU=0\\\\.5:0\\\\.95:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(bbox)/IoU=0.75\",\n \"Regex\": \".*mAP\\\\(bbox\\\\)/IoU=0\\\\.75:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(bbox)/large\",\n \"Regex\": \".*mAP\\\\(bbox\\\\)/large:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(bbox)/medium\",\n \"Regex\": \".*mAP\\\\(bbox\\\\)/medium:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(bbox)/small\",\n \"Regex\": \".*mAP\\\\(bbox\\\\)/small:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(segm)/IoU=0.5\",\n \"Regex\": \".*mAP\\\\(segm\\\\)/IoU=0\\\\.5:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(segm)/IoU=0.5:0.95\",\n \"Regex\": \".*mAP\\\\(segm\\\\)/IoU=0\\\\.5:0\\\\.95:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(segm)/IoU=0.75\",\n \"Regex\": \".*mAP\\\\(segm\\\\)/IoU=0\\\\.75:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(segm)/large\",\n \"Regex\": \".*mAP\\\\(segm\\\\)/large:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(segm)/medium\",\n \"Regex\": \".*mAP\\\\(segm\\\\)/medium:\\\\s*(\\\\S+).*\"\n },\n {\n \"Name\": \"mAP(segm)/small\",\n \"Regex\": \".*mAP\\\\(segm\\\\)/small:\\\\s*(\\\\S+).*\"\n } \n \n ]",
"_____no_output_____"
]
],
[
[
"## Define SageMaker Training Job\n\nNext, we use SageMaker [Tensorflow](https://sagemaker.readthedocs.io/en/stable/frameworks/tensorflow/sagemaker.tensorflow.html) API to define a SageMaker Training Job that uses SageMaker script mode.",
"_____no_output_____"
],
[
"### Select script\n\nIn script-mode, first we have to select an entry point script that acts as interface with SageMaker and launches the training job. For training [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) model, set ```script``` to ```\"tensorpack-mask-rcnn.py\"```. For training [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) model, set ```script``` to ```\"aws-mask-rcnn.py\"```.",
"_____no_output_____"
]
],
[
[
"script= # \"tensorpack-mask-rcnn.py\" or \"aws-mask-rcnn.py\"",
"_____no_output_____"
]
],
[
[
"### Select distribution mode\n\nWe use Message Passing Interface (MPI) to distribute the training job across multiple hosts. The ```custom_mpi_options``` below is only used by [AWS Samples Mask R-CNN](https://github.com/aws-samples/mask-rcnn-tensorflow) model, and can be safely commented out for [TensorPack Faster-RCNN/Mask-RCNN](https://github.com/tensorpack/tensorpack/tree/master/examples/FasterRCNN) model.",
"_____no_output_____"
]
],
[
[
"mpi_distribution={'mpi': \n { \n 'enabled': True,\n \"custom_mpi_options\" : \"-x TENSORPACK_FP16=1 \"\n }\n }",
"_____no_output_____"
]
],
[
[
"### Define SageMaker Tensorflow Estimator\nWe recommned using 32 GPUs, so we set ```instance_count=4``` and ```instance_type='ml.p3.16xlarge'```, because there are 8 Tesla V100 GPUs per ```ml.p3.16xlarge``` instance. We recommend using 100 GB [Amazon EBS](https://aws.amazon.com/ebs/) storage volume with each training instance, so we set ```volume_size = 100```. \n\nWe run the training job in your private VPC, so we need to set the ```subnets``` and ```security_group_ids``` prior to running the cell below. You may specify multiple subnet ids in the ```subnets``` list. The subnets included in the ```sunbets``` list must be part of the output of ```./stack-sm.sh``` CloudFormation stack script used to create this notebook instance. Specify only one security group id in ```security_group_ids``` list. The security group id must be part of the output of ```./stack-sm.sh``` script.\n\nFor ```instance_type``` below, you have the option to use ```ml.p3.16xlarge``` with 16 GB per-GPU memory and 25 Gbs network interconnectivity, or ```ml.p3dn.24xlarge``` with 32 GB per-GPU memory and 100 Gbs network interconnectivity. The ```ml.p3dn.24xlarge``` instance type offers significantly better performance than ```ml.p3.16xlarge``` for Mask R-CNN distributed TensorFlow training.\n\nWe use MPI to distribute the training job across multiple hosts.",
"_____no_output_____"
]
],
[
[
"# Give Amazon SageMaker Training Jobs Access to FileSystem Resources in Your Amazon VPC.\nsecurity_group_ids = # ['sg-xxxxxxxx'] \nsubnets = # [ 'subnet-xxxxxxx']\nsagemaker_session = sagemaker.session.Session(boto_session=session)\n\nmask_rcnn_estimator = TensorFlow(image_uri=training_image,\n role=role, \n py_version='py3',\n instance_count=4, \n instance_type='ml.p3.16xlarge',\n distribution=mpi_distribution,\n entry_point=script,\n volume_size = 100,\n max_run = 400000,\n output_path=s3_output_location,\n sagemaker_session=sagemaker_session, \n hyperparameters = hyperparameters,\n metric_definitions = metric_definitions,\n subnets=subnets,\n security_group_ids=security_group_ids)\n\n",
"_____no_output_____"
]
],
[
[
"### Launch training job\nFinally, we launch the SageMaker training job. See ```Training Jobs``` in SageMaker console to monitor the training job. ",
"_____no_output_____"
]
],
[
[
"import time\n\njob_name=f'mask-rcnn-efs-script-mode-{int(time.time())}'\nprint(f\"Launching Training Job: {job_name}\")\n\n# set wait=True below if you want to print logs in cell output\nmask_rcnn_estimator.fit(inputs=data_channels, job_name=job_name, logs=\"All\", wait=False)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b409444dd2e26b48cea5d6ed1f81aef65c56b3 | 10,678 | ipynb | Jupyter Notebook | QEC_BitFlipCode/QEC_BitFlipCode.ipynb | HectorGBoissier/QuantumKatas | 6c11c77542b2e103ef05553968cdb0107184c02d | [
"MIT"
] | 2,514 | 2019-05-06T21:55:03.000Z | 2022-03-30T20:35:50.000Z | QEC_BitFlipCode/QEC_BitFlipCode.ipynb | HectorGBoissier/QuantumKatas | 6c11c77542b2e103ef05553968cdb0107184c02d | [
"MIT"
] | 338 | 2019-05-08T22:51:25.000Z | 2022-03-31T01:56:29.000Z | QEC_BitFlipCode/QEC_BitFlipCode.ipynb | HectorGBoissier/QuantumKatas | 6c11c77542b2e103ef05553968cdb0107184c02d | [
"MIT"
] | 970 | 2019-05-07T01:18:07.000Z | 2022-03-31T04:30:53.000Z | 41.387597 | 545 | 0.567616 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b411ff112eb331ff158a3a1c97101315838751 | 10,332 | ipynb | Jupyter Notebook | python-for-data/.ipynb_checkpoints/Ex03 - Booleans and Conditionals-checkpoint.ipynb | cnhhoang850/atom-assignments | 1b792660c3113ca09efd254289b089fc52928344 | [
"MIT"
] | null | null | null | python-for-data/.ipynb_checkpoints/Ex03 - Booleans and Conditionals-checkpoint.ipynb | cnhhoang850/atom-assignments | 1b792660c3113ca09efd254289b089fc52928344 | [
"MIT"
] | null | null | null | python-for-data/.ipynb_checkpoints/Ex03 - Booleans and Conditionals-checkpoint.ipynb | cnhhoang850/atom-assignments | 1b792660c3113ca09efd254289b089fc52928344 | [
"MIT"
] | null | null | null | 29.689655 | 270 | 0.598142 | [
[
[
"# Exercise 03 - Booleans and Conditionals",
"_____no_output_____"
],
[
"## 1. Simple Function with Conditionals\n\nMany programming languages have [sign](https://en.wikipedia.org/wiki/Sign_function) available as a built-in function. Python does not, but we can define our own!\n\nIn the cell below, define a function called `sign` which takes a numerical argument and returns -1 if it's negative, 1 if it's positive, and 0 if it's 0.",
"_____no_output_____"
]
],
[
[
"# Your code goes here. Define a function called 'sign'\ndef sign(num) {\n return num % 1\n}",
"_____no_output_____"
]
],
[
[
"## 2. Singular vs Plural Nouns\n\nWe've decided to add \"print\" to our `to_smash` function from Exercise 02",
"_____no_output_____"
]
],
[
[
"def to_smash(total_candies):\n \"\"\"Return the number of leftover candies that must be smashed after distributing\n the given number of candies evenly between 3 friends.\n \n >>> to_smash(91)\n 1\n \"\"\"\n print(\"Splitting\", total_candies, \"candies\")\n return total_candies % 3\n\nto_smash(91)",
"_____no_output_____"
]
],
[
[
"What happens if we call it with `total_candies = 1`?",
"_____no_output_____"
]
],
[
[
"to_smash(1)",
"_____no_output_____"
]
],
[
[
"**Wrong grammar there!**\n\nModify the definition in the cell below to correct the grammar of our print statement.\n\n**Your Task:**\n> If there's only one candy, we should use the singular \"candy\" instead of the plural \"candies\"",
"_____no_output_____"
]
],
[
[
"def to_smash(total_candies):\n \"\"\"Return the number of leftover candies that must be smashed after distributing\n the given number of candies evenly between 3 friends.\n \n >>> to_smash(91)\n 1\n \"\"\"\n print(\"Splitting\", total_candies, \"candies\")\n return total_candies % 3\n\nto_smash(91)\nto_smash(1)",
"_____no_output_____"
]
],
[
[
"## 3. Checking weather again\n\nIn the main lesson we talked about deciding whether we're prepared for the weather. I said that I'm safe from today's weather if...\n- I have an umbrella...\n- or if the rain isn't too heavy and I have a hood...\n- otherwise, I'm still fine unless it's raining *and* it's a workday\n\nThe function below uses our first attempt at turning this logic into a Python expression. I claimed that there was a bug in that code. Can you find it?\n\nTo prove that `prepared_for_weather` is buggy, come up with a set of inputs where it returns the wrong answer.",
"_____no_output_____"
]
],
[
[
"def prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday):\n # Don't change this code. Our goal is just to find the bug, not fix it!\n return have_umbrella or rain_level < 5 and have_hood or not rain_level > 0 and is_workday\n\n# Change the values of these inputs so they represent a case where prepared_for_weather\n# returns the wrong answer.\nhave_umbrella = True\nrain_level = 0.0\nhave_hood = True\nis_workday = True\n\n# Check what the function returns given the current values of the variables above\nactual = prepared_for_weather(have_umbrella, rain_level, have_hood, is_workday)\nprint(actual)",
"_____no_output_____"
]
],
[
[
"## 4. Start being lazy...\n\nThe function `is_negative` below is implemented correctly \n- It returns True if the given number is negative and False otherwise.\n\nHowever, it's more verbose than it needs to be. We can actually reduce the number of lines of code in this function by *75%* while keeping the same behaviour. \n\n**Your task:**\n> See if you can come up with an equivalent body that uses just **one line** of code, and put it in the function `concise_is_negative`. (HINT: you don't even need Python's ternary syntax)",
"_____no_output_____"
]
],
[
[
"def is_negative(number):\n if number < 0:\n return True\n else:\n return False\n\ndef concise_is_negative(number):\n pass # Your code goes here (try to keep it to one line!)\n",
"_____no_output_____"
]
],
[
[
"## 5. Adding Toppings\n\nThe boolean variables `ketchup`, `mustard` and `onion` represent whether a customer wants a particular topping on their hot dog. We want to implement a number of boolean functions that correspond to some yes-or-no questions about the customer's order. For example:",
"_____no_output_____"
]
],
[
[
"def onionless(ketchup, mustard, onion):\n \"\"\"Return whether the customer doesn't want onions.\n \"\"\"\n return not onion",
"_____no_output_____"
]
],
[
[
"**Your task:**\n> For each of the remaining functions, fill in the body to match the English description in the docstring. ",
"_____no_output_____"
]
],
[
[
"def wants_all_toppings(ketchup, mustard, onion):\n \"\"\"Return whether the customer wants \"the works\" (all 3 toppings)\n \"\"\"\n pass\n",
"_____no_output_____"
],
[
"def wants_plain_hotdog(ketchup, mustard, onion):\n \"\"\"Return whether the customer wants a plain hot dog with no toppings.\n \"\"\"\n pass\n",
"_____no_output_____"
],
[
"def exactly_one_sauce(ketchup, mustard, onion):\n \"\"\"Return whether the customer wants either ketchup or mustard, but not both.\n (You may be familiar with this operation under the name \"exclusive or\")\n \"\"\"\n pass\n",
"_____no_output_____"
]
],
[
[
"## 6. <span title=\"A bit spicy\" style=\"color: darkgreen \">🌶️</span>\n\nWe’ve seen that calling `bool()` on an integer returns `False` if it’s equal to 0 and `True` otherwise. What happens if we call `int()` on a bool? Try it out in the notebook cell below.\n\nCan you take advantage of this to write a succinct function that corresponds to the English sentence \"*Does the customer want exactly one topping?*\"?\n\n> *HINT*: You may have already found that `int(True)` is `1`, and `int(False)` is `0`. Think about what kinds of basic arithmetic operations you might want to perform on ketchup, mustard, and onion after converting them to integers.",
"_____no_output_____"
]
],
[
[
"def exactly_one_topping(ketchup, mustard, onion):\n \"\"\"Return whether the customer wants exactly one of the three available toppings\n on their hot dog.\n \"\"\"\n pass\n",
"_____no_output_____"
]
],
[
[
"# Keep Going 💪",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b4218053e089b8489ba0c3cdebf97497c4e33d | 8,648 | ipynb | Jupyter Notebook | docs/tutorials/optimizers_lazyadam.ipynb | failure-to-thrive/addons | 63c82e318e68b07eb1162d1ff247fe9f4d3194fc | [
"Apache-2.0"
] | null | null | null | docs/tutorials/optimizers_lazyadam.ipynb | failure-to-thrive/addons | 63c82e318e68b07eb1162d1ff247fe9f4d3194fc | [
"Apache-2.0"
] | null | null | null | docs/tutorials/optimizers_lazyadam.ipynb | failure-to-thrive/addons | 63c82e318e68b07eb1162d1ff247fe9f4d3194fc | [
"Apache-2.0"
] | null | null | null | 26.609231 | 247 | 0.569958 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b439e684e20e4e8fe9a2b346c040caa68f6dea | 78,187 | ipynb | Jupyter Notebook | Tutorial/Day 5 Optimal Mind Control/Day 5.ipynb | technosap/PSST | 76bf7e74cdb457bee65235cde93b88041130fe65 | [
"MIT"
] | 5 | 2019-06-11T12:51:27.000Z | 2020-07-06T10:09:00.000Z | Tutorial/Day 5 Optimal Mind Control/Day 5.ipynb | technosap/PSST | 76bf7e74cdb457bee65235cde93b88041130fe65 | [
"MIT"
] | null | null | null | Tutorial/Day 5 Optimal Mind Control/Day 5.ipynb | technosap/PSST | 76bf7e74cdb457bee65235cde93b88041130fe65 | [
"MIT"
] | 4 | 2019-05-21T05:11:20.000Z | 2019-12-19T05:20:13.000Z | 89.766935 | 18,768 | 0.750432 | [
[
[
"<a href=\"https://colab.research.google.com/github/neurorishika/PSST/blob/master/Tutorial/Day%205%20Optimal%20Mind%20Control/Day%205.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a> <a href=\"https://kaggle.com/kernels/welcome?src=https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/Day%205.ipynb\" target=\"_parent\"><img src=\"https://kaggle.com/static/images/open-in-kaggle.svg\" alt=\"Open in Kaggle\"/></a>",
"_____no_output_____"
],
[
"## Day 5: Optimal Mind Control\n\nWelcome to Day 5! Now that we can simulate a model network of conductance-based neurons, we discuss the limitations of our approach and attempts to work around these issues.",
"_____no_output_____"
],
[
"### Memory Management\n\nUsing Python and TensorFlow allowed us to write code that is readable, parallizable and scalable across a variety of computational devices. However, our implementation is very memory intensive. The iterators in TensorFlow do not follow the normal process of memory allocation and garbage collection. Since, TensorFlow is designed to work on diverse hardware like GPUs, TPUs and distributed platforms, memory allocation is done adaptively during the TensorFlow session and not cleared until the Python kernel has stopped execution. The memory used increases linearly with time as the state matrix is computed recursively by the tf.scan function. The maximum memory used by the computational graph is 2 times the total state matrix size at the point when the computation finishes and copies the final data into the memory. The larger the network and longer the simulation, the larger the solution matrix. Each run is limited by the total available memory. For a system with a limited memory of K bytes, The length of a given simulation (L timesteps) of a given network (N differential equations) with 64-bit floating-point precision will follow:\n\n$$2\\times64\\times L\\times N=K$$ \n\nThat is, for any given network, our maximum simulation length is limited. One way to improve our maximum length is to divide the simulation into smaller batches. There will be a small queuing time between batches, which will slow down our code by a small amount but we will be able to simulate longer times. Thus, if we split the simulation into K sequential batches, the maximum memory for the simulation becomes $(1+\\frac{1}{K})$ times the total matrix size. Thus the memory relation becomes: \n\n$$\\Big(1+\\frac{1}{K}\\Big)\\times64\\times L\\times N=K$$ \n\nThis way, we can maximize the length of out simulation that we can run in a single python kernel.\n\nLet us implement this batch system for our 3 neuron feed-forward model.\n\n### Implementing the Model\n\nTo improve the readability of our code we separate the integrator into a independent import module. The integrator code was placed in a file called tf integrator.py. The file must be present in the same directory as the implementation of the model.\n\nNote: If you are using Jupyter Notebook, remember to remove the %matplotlib inline command as it is specific to jupyter.\n\n#### Importing tf_integrator and other requirements\n\nOnce the Integrator is saved in tf_integrator.py in the same directory as the Notebook, we can start importing the essentials including the integrator.",
"_____no_output_____"
],
[
"**WARNING: If you are running this notebook using Kaggle, make sure you have logged in to your verified Kaggle account and enabled Internet Access for the kernel. For instructions on enabling Internet on Kaggle Kernels, visit: https://www.kaggle.com/product-feedback/63544**",
"_____no_output_____"
]
],
[
[
"#@markdown Import required files and code from previous tutorials\n\n!wget --no-check-certificate \\\n \"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/tf_integrator.py\" \\\n -O \"tf_integrator.py\"\n!wget --no-check-certificate \\\n \"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/call.py\" \\\n -O \"call.py\"\n!wget --no-check-certificate \\\n \"https://raw.githubusercontent.com/neurorishika/PSST/master/Tutorial/Day%205%20Optimal%20Mind%20Control/run.py\" \\\n -O \"run.py\"",
"_____no_output_____"
],
[
"import numpy as np\nimport tf_integrator as tf_int\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport tensorflow.compat.v1 as tf\ntf.disable_eager_execution()",
"_____no_output_____"
]
],
[
[
"### Recall the Model\n\nFor implementing a Batch system, we do not need to change how we construct our model only how we execute it.\n\n#### Step 1: Initialize Parameters and Dynamical Equations; Define Input",
"_____no_output_____"
]
],
[
[
"n_n = 3 # Number of simultaneous neurons to simulate\n\nsim_res = 0.01 # Time Resolution of the Simulation\nsim_time = 700 # Length of the Simulation\nt = np.arange(0,sim_time,sim_res) # Time points at which to simulate the network\n\n# Acetylcholine\n\nach_mat = np.zeros((n_n,n_n)) # Ach Synapse Connectivity Matrix\nach_mat[1,0]=1\n\n## PARAMETERS FOR ACETYLCHLOLINE SYNAPSES ##\n\nn_ach = int(np.sum(ach_mat)) # Number of Acetylcholine (Ach) Synapses \nalp_ach = [10.0]*n_ach # Alpha for Ach Synapse\nbet_ach = [0.2]*n_ach # Beta for Ach Synapse\nt_max = 0.3 # Maximum Time for Synapse\nt_delay = 0 # Axonal Transmission Delay\nA = [0.5]*n_n # Synaptic Response Strength\ng_ach = [0.35]*n_n # Ach Conductance\nE_ach = [0.0]*n_n # Ach Potential\n\n# GABAa\n\ngaba_mat = np.zeros((n_n,n_n)) # GABAa Synapse Connectivity Matrix\ngaba_mat[2,1] = 1\n\n## PARAMETERS FOR GABAa SYNAPSES ##\n\nn_gaba = int(np.sum(gaba_mat)) # Number of GABAa Synapses\nalp_gaba = [10.0]*n_gaba # Alpha for GABAa Synapse\nbet_gaba = [0.16]*n_gaba # Beta for GABAa Synapse\nV0 = [-20.0]*n_n # Decay Potential\nsigma = [1.5]*n_n # Decay Time Constant\ng_gaba = [0.8]*n_n # fGABA Conductance\nE_gaba = [-70.0]*n_n # fGABA Potential\n\n## Storing Firing Thresholds ##\nF_b = [0.0]*n_n # Fire threshold\n\ndef I_inj_t(t):\n \"\"\"\n This function returns the external current to be injected into the network at any time step from the current_input matrix.\n\n Parameters:\n -----------\n t: float\n The time at which the current injection is being performed.\n \"\"\"\n # Turn indices to integer and extract from matrix\n index = tf.cast(t/sim_res,tf.int32)\n return tf.constant(current_input.T,dtype=tf.float64)[index] \n\n## Acetylcholine Synaptic Current ##\n\ndef I_ach(o,V):\n \"\"\"\n This function returns the synaptic current for the Acetylcholine (Ach) synapses for each neuron.\n\n Parameters:\n -----------\n o: float\n The fraction of open acetylcholine channels for each synapse.\n V: float\n The membrane potential of the postsynaptic neuron.\n \"\"\"\n o_ = tf.constant([0.0]*n_n**2,dtype=tf.float64) # Initialize the flattened matrix to store the synaptic open fractions\n ind = tf.boolean_mask(tf.range(n_n**2),ach_mat.reshape(-1) == 1) # Get the indices of the synapses that exist\n o_ = tf.tensor_scatter_nd_update(o_,tf.reshape(ind,[-1,1]),o) # Update the flattened open fraction matrix\n o_ = tf.transpose(tf.reshape(o_,(n_n,n_n))) # Reshape and Transpose the matrix to be able to multiply it with the conductance matrix\n return tf.reduce_sum(tf.transpose((o_*(V-E_ach))*g_ach),1) # Calculate the synaptic current\n\n## GABAa Synaptic Current ##\n\ndef I_gaba(o,V):\n \"\"\"\n This function returns the synaptic current for the GABA synapses for each neuron.\n\n Parameters:\n -----------\n o: float\n The fraction of open GABA channels for each synapse.\n V: float\n The membrane potential of the postsynaptic neuron.\n \"\"\"\n o_ = tf.constant([0.0]*n_n**2,dtype=tf.float64) # Initialize the flattened matrix to store the synaptic open fractions\n ind = tf.boolean_mask(tf.range(n_n**2),gaba_mat.reshape(-1) == 1) # Get the indices of the synapses that exist\n o_ = tf.tensor_scatter_nd_update(o_,tf.reshape(ind,[-1,1]),o) # Update the flattened open fraction matrix\n o_ = tf.transpose(tf.reshape(o_,(n_n,n_n))) # Reshape and Transpose the matrix to be able to multiply it with the conductance matrix\n return tf.reduce_sum(tf.transpose((o_*(V-E_gaba))*g_gaba),1) # Calculate the synaptic current\n\n## Other Currents ##\n\ndef I_K(V, n):\n \"\"\"\n This function determines the K-channel current.\n\n Parameters:\n -----------\n V: float\n The membrane potential.\n n: float \n The K-channel gating variable n.\n \"\"\"\n return g_K * n**4 * (V - E_K)\n\ndef I_Na(V, m, h):\n \"\"\"\n This function determines the Na-channel current.\n \n Parameters:\n -----------\n V: float\n The membrane potential.\n m: float\n The Na-channel gating variable m.\n h: float\n The Na-channel gating variable h.\n \"\"\"\n return g_Na * m**3 * h * (V - E_Na)\n\ndef I_L(V):\n \"\"\"\n This function determines the leak current.\n\n Parameters:\n -----------\n V: float\n The membrane potential.\n \"\"\"\n return g_L * (V - E_L)\n\ndef dXdt(X, t):\n \"\"\"\n This function determines the derivatives of the membrane voltage and gating variables for n_n neurons.\n\n Parameters:\n -----------\n X: float\n The state vector given by the [V1,V2,...,Vn_n,m1,m2,...,mn_n,h1,h2,...,hn_n,n1,n2,...,nn_n] where \n Vx is the membrane potential for neuron x\n mx is the Na-channel gating variable for neuron x \n hx is the Na-channel gating variable for neuron x\n nx is the K-channel gating variable for neuron x.\n t: float\n The time points at which the derivatives are being evaluated.\n \"\"\"\n V = X[:1*n_n] # First n_n values are Membrane Voltage\n m = X[1*n_n:2*n_n] # Next n_n values are Sodium Activation Gating Variables\n h = X[2*n_n:3*n_n] # Next n_n values are Sodium Inactivation Gating Variables\n n = X[3*n_n:4*n_n] # Next n_n values are Potassium Gating Variables\n o_ach = X[4*n_n : 4*n_n + n_ach] # Next n_ach values are Acetylcholine Synapse Open Fractions\n o_gaba = X[4*n_n + n_ach : 4*n_n + n_ach + n_gaba] # Next n_gaba values are GABAa Synapse Open Fractions\n fire_t = X[-n_n:] # Last n_n values are the last fire times as updated by the modified integrator\n \n dVdt = (I_inj_t(t) - I_Na(V, m, h) - I_K(V, n) - I_L(V) - I_ach(o_ach,V) - I_gaba(o_gaba,V)) / C_m # The derivative of the membrane potential\n \n ## Updation for gating variables ##\n \n m0,tm,h0,th = Na_prop(V) # Calculate the dynamics of the Na-channel gating variables for all n_n neurons\n n0,tn = K_prop(V) # Calculate the dynamics of the K-channel gating variables for all n_n neurons\n\n dmdt = - (1.0/tm)*(m-m0) # The derivative of the Na-channel gating variable m for all n_n neurons\n dhdt = - (1.0/th)*(h-h0) # The derivative of the Na-channel gating variable h for all n_n neurons\n dndt = - (1.0/tn)*(n-n0) # The derivative of the K-channel gating variable n for all n_n neurons\n \n ## Updation for o_ach ##\n \n A_ = tf.constant(A,dtype=tf.float64) # Get the synaptic response strengths of the pre-synaptic neurons\n Z_ = tf.zeros(tf.shape(A_),dtype=tf.float64) # Create a zero matrix of the same size as A_\n \n T_ach = tf.where(tf.logical_and(tf.greater(t,fire_t+t_delay),tf.less(t,fire_t+t_max+t_delay)),A_,Z_) # Find which synapses would have received an presynaptic spike in the past window and assign them the corresponding synaptic response strength\n T_ach = tf.multiply(tf.constant(ach_mat,dtype=tf.float64),T_ach) # Find the postsynaptic neurons that would have received an presynaptic spike in the past window\n T_ach = tf.boolean_mask(tf.reshape(T_ach,(-1,)),ach_mat.reshape(-1) == 1) # Get the pre-synaptic activation function for only the existing synapses\n \n do_achdt = alp_ach*(1.0-o_ach)*T_ach - bet_ach*o_ach # Calculate the derivative of the open fraction of the acetylcholine synapses\n \n ## Updation for o_gaba ##\n \n T_gaba = 1.0/(1.0+tf.exp(-(V-V0)/sigma)) # Calculate the presynaptic activation function for all n_n neurons\n T_gaba = tf.multiply(tf.constant(gaba_mat,dtype=tf.float64),T_gaba) # Find the postsynaptic neurons that would have received an presynaptic spike in the past window\n T_gaba = tf.boolean_mask(tf.reshape(T_gaba,(-1,)),gaba_mat.reshape(-1) == 1) # Get the pre-synaptic activation function for only the existing synapses\n \n do_gabadt = alp_gaba*(1.0-o_gaba)*T_gaba - bet_gaba*o_gaba # Calculate the derivative of the open fraction of the GABAa synapses\n \n ## Updation for fire times ##\n \n dfdt = tf.zeros(tf.shape(fire_t),dtype=fire_t.dtype) # zero change in fire_t as it will be updated by the modified integrator\n \n out = tf.concat([dVdt,dmdt,dhdt,dndt,do_achdt,do_gabadt,dfdt],0) # Concatenate the derivatives of the membrane potential, gating variables, and open fractions\n return out\n\n\ndef K_prop(V):\n \"\"\"\n This function determines the K-channel gating dynamics.\n\n Parameters:\n -----------\n V: float\n The membrane potential.\n \"\"\"\n T = 22 # Temperature\n phi = 3.0**((T-36.0)/10) # Temperature-correction factor\n V_ = V-(-50) # Voltage baseline shift\n \n alpha_n = 0.02*(15.0 - V_)/(tf.exp((15.0 - V_)/5.0) - 1.0) # Alpha for the K-channel gating variable n\n beta_n = 0.5*tf.exp((10.0 - V_)/40.0) # Beta for the K-channel gating variable n\n \n t_n = 1.0/((alpha_n+beta_n)*phi) # Time constant for the K-channel gating variable n\n n_0 = alpha_n/(alpha_n+beta_n) # Steady-state value for the K-channel gating variable n\n \n return n_0, t_n\n\n\ndef Na_prop(V):\n \"\"\"\n This function determines the Na-channel gating dynamics.\n \n Parameters:\n -----------\n V: float\n The membrane potential.\n \"\"\"\n T = 22 # Temperature \n phi = 3.0**((T-36)/10) # Temperature-correction factor\n V_ = V-(-50) # Voltage baseline shift\n \n alpha_m = 0.32*(13.0 - V_)/(tf.exp((13.0 - V_)/4.0) - 1.0) # Alpha for the Na-channel gating variable m\n beta_m = 0.28*(V_ - 40.0)/(tf.exp((V_ - 40.0)/5.0) - 1.0) # Beta for the Na-channel gating variable m\n \n alpha_h = 0.128*tf.exp((17.0 - V_)/18.0) # Alpha for the Na-channel gating variable h\n beta_h = 4.0/(tf.exp((40.0 - V_)/5.0) + 1.0) # Beta for the Na-channel gating variable h\n \n t_m = 1.0/((alpha_m+beta_m)*phi) # Time constant for the Na-channel gating variable m\n t_h = 1.0/((alpha_h+beta_h)*phi) # Time constant for the Na-channel gating variable h\n \n m_0 = alpha_m/(alpha_m+beta_m) # Steady-state value for the Na-channel gating variable m\n h_0 = alpha_h/(alpha_h+beta_h) # Steady-state value for the Na-channel gating variable h\n \n return m_0, t_m, h_0, t_h\n\n\n# Initializing the Parameters\n\nC_m = [1.0]*n_n # Membrane capacitances\ng_K = [10.0]*n_n # K-channel conductances\nE_K = [-95.0]*n_n # K-channel reversal potentials\n\ng_Na = [100]*n_n # Na-channel conductances\nE_Na = [50]*n_n # Na-channel reversal potentials\n\ng_L = [0.15]*n_n # Leak conductances\nE_L = [-55.0]*n_n # Leak reversal potentials\n\n# Creating the Current Input\ncurrent_input= np.zeros((n_n,t.shape[0])) # The current input to the network\ncurrent_input[0,int(100/sim_res):int(200/sim_res)] = 2.5\ncurrent_input[0,int(300/sim_res):int(400/sim_res)] = 5.0\ncurrent_input[0,int(500/sim_res):int(600/sim_res)] = 7.5",
"_____no_output_____"
]
],
[
[
"#### Step 2: Define the Initial Condition of the Network and Add some Noise to the initial conditions",
"_____no_output_____"
]
],
[
[
"# Initializing the State Vector and adding 1% noise\nstate_vector = [-71]*n_n+[0,0,0]*n_n+[0]*n_ach+[0]*n_gaba+[-9999999]*n_n\nstate_vector = np.array(state_vector)\nstate_vector = state_vector + 0.01*state_vector*np.random.normal(size=state_vector.shape)",
"_____no_output_____"
]
],
[
[
"#### Step 3: Splitting Time Series into independent batches and Run Each Batch Sequentially\n\nSince we will be dividing the computation into batches, we have to split the time array such that for each new call, the final state vector of the last batch will be the initial condition for the current batch. The function $np.array\\_split()$ splits the array into non-overlapping vectors. Therefore, we append the last time of the previous batch to the beginning of the current time array batch.",
"_____no_output_____"
]
],
[
[
"# Define the Number of Batches\nn_batch = 2\n\n# Split t array into batches using numpy\nt_batch = np.array_split(t,n_batch)\n\n# Iterate over the batches of time array\nfor n,i in enumerate(t_batch):\n \n # Inform start of Batch Computation\n print(\"Batch\",(n+1),\"Running...\",end=\"\")\n \n # In np.array_split(), the split edges are present in only one array and since \n # our initial vector to successive calls is corresposnding to the last output\n # our first element in the later time array should be the last element of the \n # previous output series, Thus, we append the last time to the beginning of \n # the current time array batch.\n if n>0:\n i = np.append(i[0]-sim_res,i)\n \n # Set state_vector as the initial condition\n init_state = tf.constant(state_vector, dtype=tf.float64)\n # Create the Integrator computation graph over the current batch of t array\n tensor_state = tf_int.odeint(dXdt, init_state, i, n_n, F_b)\n \n # Initialize variables and run session\n with tf.Session() as sess:\n tf.global_variables_initializer().run()\n state = sess.run(tensor_state)\n sess.close()\n \n # Reset state_vector as the last element of output\n state_vector = state[-1,:]\n \n # Save the output of the simulation to a binary file\n np.save(\"part_\"+str(n+1),state)\n\n # Clear output\n state=None\n \n print(\"Finished\")",
"Batch 1 Running...Finished\nBatch 2 Running...Finished\n"
]
],
[
[
"#### Putting the Output Together\n\nThe output from our batch implementation is a set of binary files that store parts of our total simulation. To get the overall output we have to stitch them back together.",
"_____no_output_____"
]
],
[
[
"overall_state = []\n\n# Iterate over the generated output files\nfor n,i in enumerate([\"part_\"+str(n+1)+\".npy\" for n in range(n_batch)]):\n \n # Since the first element in the series was the last output, we remove them\n if n>0:\n overall_state.append(np.load(i)[1:,:])\n else:\n overall_state.append(np.load(i))\n\n# Concatenate all the matrix to get a single state matrix\noverall_state = np.concatenate(overall_state)",
"_____no_output_____"
]
],
[
[
"#### Visualizing the Overall Data\nFinally, we plot the same voltage traces of the 3 neurons from Day 4 as a Voltage vs Time heatmap. While this visualization may seem unnecessary for just 3 neurons, it becomes an useful tool when on visualizes the dynamics of a large network of neurons as illustrated in the Example Implementation of the Locust Antennal Lobe. ",
"_____no_output_____"
]
],
[
[
"# Plot the voltage traces of the three neurons\nplt.figure(figsize=(12,6)) \nsns.heatmap(overall_state[::100,:3].T,xticklabels=100,yticklabels=5,cmap='RdBu_r')\nplt.xlabel(\"Time (in ms)\")\nplt.ylabel(\"Neuron Number\")\nplt.title(\"Voltage vs Time Heatmap for Projection Neurons (PNs)\")\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"By this method, we have maximized the usage of our available memory but we can go further and develop a method to allow indefinitely long simulation. The issue behind this entire algorithm is that the memory is not cleared until the python kernel finishes. One way to overcome this is to save the parameters of the model (such as connectivity matrix) and the state vector in a file, and start a new python kernel from a python script to compute successive batches. This way after each large batch, the memory gets cleaned. By combining the previous batch implementation and this system, we can maximize our computability.\n\n### Implementing a Runner and a Caller\n\nFirstly, we have to create an implementation of the model that takes in previous input as current parameters. Thus, we create a file, which we call \"run.py\" that takes an argument ie. the current batch number. The implementation for \"run.py\" is mostly same as the above model but there is a small difference.\n\nWhen the batch number is 0, we initialize all variable parameters and save them, but otherwise we use the saved values. The parameters we save include: Acetylcholine Matrix, GABAa Matrix and Final/Initial State Vector. It will also save the files with both batch number and sub-batch number listed.\n\nThe time series will be created and split initially by the caller, which we call \"call.py\", and stored in a file. Each execution of the Runner will extract its relevant time series and compute on it.",
"_____no_output_____"
],
[
"#### Implementing the Runner code\n\n\"run.py\" is essentially identical to the batch-implemented model we developed above with the changes described below:\n\n\n```\n# Additional Imports #\n\nimport sys\n\n# Duration of Simulation #\n\n# t = np.arange(0,sim_time,sim_res) \nt = np.load(\"time.npy\",allow_pickle=True)[int(sys.argv[1])] # get first argument to run.py\n\n# Connectivity Matrix Definitions #\n\nif sys.argv[1] == '0':\n ach_mat = np.zeros((n_n,n_n)) # Ach Synapse Connectivity Matrix\n ach_mat[1,0]=1 # If connectivity is random, once initialized it will be the same.\n np.save(\"ach_mat\",ach_mat)\nelse:\n ach_mat = np.load(\"ach_mat.npy\")\n \nif sys.argv[1] == '0':\n gaba_mat = np.zeros((n_n,n_n)) # GABAa Synapse Connectivity Matrix\n gaba_mat[2,1] = 1 # If connectivity is random, once initialized it will be the same.\n np.save(\"gaba_mat\",gaba_mat)\nelse:\n gaba_mat = np.load(\"gaba_mat.npy\")\n\n# Current Input Definition #\n \nif sys.argv[1] == '0':\n current_input= np.zeros((n_n,int(sim_time/sim_res)))\n current_input[0,int(100/sim_res):int(200/sim_res)] = 2.5\n current_input[0,int(300/sim_res):int(400/sim_res)] = 5.0\n current_input[0,int(500/sim_res):int(600/sim_res)] = 7.5\n np.save(\"current_input\",current_input)\nelse:\n current_input = np.load(\"current_input.npy\")\n \n# State Vector Definition #\n\nif sys.argv[1] == '0':\n state_vector = [-71]*n_n+[0,0,0]*n_n+[0]*n_ach+[0]*n_gaba+[-9999999]*n_n\n state_vector = np.array(state_vector)\n state_vector = state_vector + 0.01*state_vector*np.random.normal(size=state_vector.shape)\n np.save(\"state_vector\",state_vector)\nelse:\n state_vector = np.load(\"state_vector.npy\")\n\n# Saving of Output #\n\n# np.save(\"part_\"+str(n+1),state)\nnp.save(\"batch\"+str(int(sys.argv[1])+1)+\"_part_\"+str(n+1),state)\n```\n\n",
"_____no_output_____"
],
[
"#### Implementing the Caller code\n\nThe caller will create the time series, split it and use python subprocess module to call \"run.py\" with appropriate arguments. The code for \"call.py\" is given below.\n\n```\nfrom subprocess import call\nimport numpy as np\n\ntotal_time = 700\nn_splits = 2\ntime = np.split(np.arange(0,total_time,0.01),n_splits)\n\n# Append the last time point to the beginning of the next batch\nfor n,i in enumerate(time):\n if n>0:\n time[n] = np.append(i[0]-0.01,i)\n\nnp.save(\"time\",time)\n\n# call successive batches with a new python subprocess and pass the batch number\nfor i in range(n_splits):\n call(['python','run.py',str(i)])\n\nprint(\"Simulation Completed.\")\n```",
"_____no_output_____"
],
[
"#### Using call.py",
"_____no_output_____"
]
],
[
[
"!python call.py",
"Batch 1 Running...Finished\nBatch 2 Running...Finished"
]
],
[
[
"#### Combining all Data\n\nJust like we merged all the batches, we merge all the sub-batches and batches.",
"_____no_output_____"
]
],
[
[
"n_splits = 2\nn_batch = 2\n\noverall_state = []\n\n# Iterate over the generated output files\nfor n,i in enumerate([\"batch\"+str(x+1) for x in range(n_splits)]):\n for m,j in enumerate([\"_part_\"+str(x+1)+\".npy\" for x in range(n_batch)]):\n \n # Since the first element in the series was the last output, we remove them\n if n>0 and m>0:\n overall_state.append(np.load(i+j)[1:,:])\n else:\n overall_state.append(np.load(i+j))\n\n# Concatenate all the matrix to get a single state matrix\noverall_state = np.concatenate(overall_state)",
"_____no_output_____"
],
[
"# Plot the simulation results\nplt.figure(figsize=(12,6))\nsns.heatmap(overall_state[::100,:3].T,xticklabels=100,yticklabels=5,cmap='RdBu_r')\nplt.xlabel(\"Time (in ms)\")\nplt.ylabel(\"Neuron Number\")\nplt.title(\"Voltage vs Time Heatmap for Projection Neurons (PNs)\")\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0b43aefafe18f7481d153f38b83592a0af6d1f0 | 111,222 | ipynb | Jupyter Notebook | Module 5/bike_trippin_resample_unsolved.ipynb | KathiaF/PyBer_Analysis | 630659cc46972da3d6f3ace11115efab26e82fe2 | [
"MIT"
] | null | null | null | Module 5/bike_trippin_resample_unsolved.ipynb | KathiaF/PyBer_Analysis | 630659cc46972da3d6f3ace11115efab26e82fe2 | [
"MIT"
] | null | null | null | Module 5/bike_trippin_resample_unsolved.ipynb | KathiaF/PyBer_Analysis | 630659cc46972da3d6f3ace11115efab26e82fe2 | [
"MIT"
] | null | null | null | 74.495646 | 65,004 | 0.7103 | [
[
[
"%matplotlib notebook",
"_____no_output_____"
],
[
"# Import Dependencies\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Import our data into pandas from CSV\n#bike_trips_df = pd.read_csv(\"Resources/trip.csv\", low_memory=False)\n#bike_trips_df.head()\n\npath = \"Resources/trip.csv\"\nbike_trips_df = pd.read_csv(path, low_memory=False)\nbike_trips_df.head()",
"_____no_output_____"
],
[
"# Get the names of the columns.\nbike_trips_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 286858 entries, 0 to 286857\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 stoptime 286858 non-null object \n 1 bikeid 286858 non-null object \n 2 tripduration 286858 non-null float64\n 3 from_station_name 286858 non-null object \n 4 to_station_name 286858 non-null object \n 5 from_station_id 286858 non-null object \n 6 to_station_id 286858 non-null object \n 7 usertype 286858 non-null object \n 8 gender 181558 non-null object \n 9 birthyear 181554 non-null object \ndtypes: float64(1), object(9)\nmemory usage: 21.9+ MB\n"
],
[
"#create a clean dataframe after dropping the null values.\nclean_bike_trips_df = bike_trips_df.dropna()\n",
"_____no_output_____"
],
[
"#check for null values again\nclean_bike_trips_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 181554 entries, 0 to 286849\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 stoptime 181554 non-null object \n 1 bikeid 181554 non-null object \n 2 tripduration 181554 non-null float64\n 3 from_station_name 181554 non-null object \n 4 to_station_name 181554 non-null object \n 5 from_station_id 181554 non-null object \n 6 to_station_id 181554 non-null object \n 7 usertype 181554 non-null object \n 8 gender 181554 non-null object \n 9 birthyear 181554 non-null object \ndtypes: float64(1), object(9)\nmemory usage: 15.2+ MB\n"
],
[
"# Split up the data into groups based upon 'gender' and 'stoptime'\n# And, find out how many bike trips each gender took.\ngender_stoptime = clean_bike_trips_df.groupby([\"gender\", \"stoptime\"]).count(['tripduration'])\n",
"_____no_output_____"
],
[
"# Reset the index of the previous Pandas Series to convert to a DataFrame. \ngender_stoptime['stoptime'] = \n",
"_____no_output_____"
],
[
"# Get the datatypes for the DataFrame columns.\ngender_stoptime.dtypes",
"_____no_output_____"
],
[
"# Convert the 'stoptime' column to a datetime object.\n",
"_____no_output_____"
],
[
"# Check the datatypes for each column.\n",
"_____no_output_____"
],
[
"# Check the DataFrame.\n",
"_____no_output_____"
],
[
"# Create a pivot table with the 'stoptime' as the index and the columns ='gender' with the trip counts in each row.\n",
"_____no_output_____"
],
[
"# Drop the stoptime column.\n",
"_____no_output_____"
],
[
"# Create a new DataFrame from the pivot table DataFrame by filtering for the given dates, '2015-01-01':'2015-12-31'. \n",
"_____no_output_____"
],
[
"# Resample the DataFrame by the week. i.e., \"W\", and get the trip counts for each week. \n",
"_____no_output_____"
],
[
"# Plot the resample DataFrame \n\n# Add a title \n\n# Add a x- and y-axis label.\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b4461a379f82e4eaaefc8e84c0b61caaeae22a | 409,910 | ipynb | Jupyter Notebook | doc/getting_started/2_wavefronts_optical_systems.ipynb | rahulbhadani/hcipy | b52726cb9502b5225ddff9d7b1ff417f2350cda8 | [
"MIT"
] | null | null | null | doc/getting_started/2_wavefronts_optical_systems.ipynb | rahulbhadani/hcipy | b52726cb9502b5225ddff9d7b1ff417f2350cda8 | [
"MIT"
] | null | null | null | doc/getting_started/2_wavefronts_optical_systems.ipynb | rahulbhadani/hcipy | b52726cb9502b5225ddff9d7b1ff417f2350cda8 | [
"MIT"
] | null | null | null | 1,601.210938 | 152,248 | 0.960899 | [
[
[
"# II - Wavefronts and optical systems\n\nFirst let's import HCIPy, and a few supporting libraries:",
"_____no_output_____"
]
],
[
[
"from hcipy import *\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Wavefronts in HCIPy are monochromatic. They consist of an electric field (as an HCIP `Field`), and a wavelength. If broadband images are needed, multiple `Wavefront`s must be constructed and propagated through the optical system, sampling the required wavelength range. Let us construct a `Wavefront`.",
"_____no_output_____"
]
],
[
[
"pupil_grid = make_pupil_grid(1024)\naperture = circular_aperture(1)(pupil_grid)\n\nwavefront = Wavefront(aperture, 1)",
"_____no_output_____"
]
],
[
[
"A note must be made at this point regarding units. HCIPy is averse w.r.t. the used units. If the user fills in all quantities in SI, then all calculations in HCIPy will be returned in SI units. This allows the user to use any unit he/she wants, while still being able to seamlessly use dimensionless quantities. Ie. the convention that is used in this document, is that if the diameter of the aperture is 1, the wavelength is 1, and the focal length is 1 as well, then the focal-plane will be given in $\\lambda/D$.\n\nTo propagate this wavefront to the focal plane, we first need to construct a grid on which the focal plane will be sampled:",
"_____no_output_____"
]
],
[
[
"focal_grid = make_focal_grid(pupil_grid, 8, 16)",
"_____no_output_____"
]
],
[
[
"This constructs a `Grid` with 8 samples per $\\lambda/D$ and a 16 $\\lambda/D$ radius field of view (so 32 $\\lambda/D$ diameter field of view). Now we can construct a Fraunhofer propagator that can actually propagate the light to the focal plane.",
"_____no_output_____"
]
],
[
[
"prop = FraunhoferPropagator(pupil_grid, focal_grid)\n\nimg = prop.forward(wavefront)\n\nimshow_field(np.log10(img.intensity / img.intensity.max()), vmin=-5)\nplt.colorbar()\nplt.show()",
"_____no_output_____"
]
],
[
[
"All Fourier transforms concerning the propagation are done internally. In this case a Matrix Fourier transform was used, as it was deemed quicker than a Fast Fourier transform in this case. Also note that when defining the propagator, we didn't pass the wavelength of the wavefront. This wavelength is taken from the `Wavefront` object during the propagation.\n\nAlso note that a `Wavefront` supports many properties to make it easier to use. One that we used above is `Wavefront.intensity`, but others exist as well: for example `Wavefront.phase` and `Wavefront.amplitude`, which yield the phase and amplitude of the electric field respectively. All these properties are returned as `Field`s, and can therefore be shown using `imshow_field()`.\n\nFor a more interesting results, let's do a propagation with physical quantities. We calculate the intensity pattern of a circular aperture with a diameter of 1cm, after a free-space propagation of 2m, at a wavelength of 500nm.",
"_____no_output_____"
]
],
[
[
"pupil_grid_2 = make_pupil_grid(1024, 0.015)\naperture_2 = circular_aperture(0.01)(pupil_grid_2)\n\nfresnel_prop = FresnelPropagator(pupil_grid_2, 2)\n\nwf = Wavefront(aperture_2, 500e-9)\nimg = fresnel_prop(wf)\n\nimshow_field(img.intensity)\nplt.show()",
"_____no_output_____"
]
],
[
[
"The propagators shown previously are part of a larger group of optical elements. All `OpticalElement`s can propagate a `Wavefront` through them. Examples include simple `Apodizer`s, which act as an infinitely-thin screen with a (complex) transmission. A little more complicated example is `SurfaceAberration`, which simulates a surface error with a power-law PSD (power spectral density). Optical elements can be linked to represent more complicated optical systems.",
"_____no_output_____"
]
],
[
[
"aberration = SurfaceAberration(pupil_grid, 0.25, 1)\n\nwf = Wavefront(aperture)\nimg = prop(aberration(wf))\n\nimshow_field(np.log10(img.intensity / img.intensity.max()), vmin=-5)\nplt.colorbar()\nplt.show()",
"c:\\users\\emiel por\\documents\\github\\hcipy\\hcipy\\optics\\aberration.py:39: RuntimeWarning: divide by zero encountered in power\n res = Field(grid.as_('polar').r**exponent, grid)\n"
]
],
[
[
"These simple optical elements can be combined into more complicated optical systems. These include full wavefront sensor implementations and coronagraphs. Both of these will be handled in later sections.\n\nTo convert a `Wavefront` into an observed image, one can simply use the `Wavefront.power` attribute, which is the `Wavefront.intensity` multiplied by the weight at each pixel. If one wants to use a more complicated detector model, HCIPy supplies a `Detector` class. and its derivatives. A detector uses an integration/readout scheme:",
"_____no_output_____"
]
],
[
[
"flat_field = 0.01\ndark = 10\ndetector = NoisyDetector(focal_grid, dark_current_rate=dark, flat_field=flat_field)\n\nwf.total_power = 5000\nimg = prop(aberration(wf))\n\ndetector.integrate(img, 0.5)\nimage = detector.read_out()\n\nimshow_field(np.log10(image), vmax=np.log10(image).max(), vmin=0)\nplt.colorbar()\nplt.show()",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:11: RuntimeWarning: divide by zero encountered in log10\n # This is added back by InteractiveShellApp.init_path()\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b44b362f24fac5d37537a5121aa9636dd3d125 | 28,527 | ipynb | Jupyter Notebook | notebooks/M6-ensemble_ex_03.ipynb | datagistips/scikit-learn-mooc | 9eb67c53173218b5cd3061712c827c6a663e425a | [
"CC-BY-4.0"
] | 1 | 2021-07-14T09:41:21.000Z | 2021-07-14T09:41:21.000Z | notebooks/M6-ensemble_ex_03.ipynb | datagistips/scikit-learn-mooc | 9eb67c53173218b5cd3061712c827c6a663e425a | [
"CC-BY-4.0"
] | null | null | null | notebooks/M6-ensemble_ex_03.ipynb | datagistips/scikit-learn-mooc | 9eb67c53173218b5cd3061712c827c6a663e425a | [
"CC-BY-4.0"
] | null | null | null | 135.199052 | 22,676 | 0.889508 | [
[
[
"# 📝 Exercise M6.03\n\nThis exercise aims at verifying if AdaBoost can over-fit.\nWe will make a grid-search and check the scores by varying the\nnumber of estimators.\n\nWe will first load the California housing dataset and split it into a\ntraining and a testing set.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import fetch_california_housing\nfrom sklearn.model_selection import train_test_split\n\ndata, target = fetch_california_housing(return_X_y=True, as_frame=True)\ntarget *= 100 # rescale the target in k$\ndata_train, data_test, target_train, target_test = train_test_split(\n data, target, random_state=0, test_size=0.5)",
"_____no_output_____"
]
],
[
[
"<div class=\"admonition note alert alert-info\">\n<p class=\"first admonition-title\" style=\"font-weight: bold;\">Note</p>\n<p class=\"last\">If you want a deeper overview regarding this dataset, you can refer to the\nAppendix - Datasets description section at the end of this MOOC.</p>\n</div>",
"_____no_output_____"
],
[
"Then, create an `AdaBoostRegressor`. Use the function\n`sklearn.model_selection.validation_curve` to get training and test scores\nby varying the number of estimators. Use the mean absolute error as a metric\nby passing `scoring=\"neg_mean_absolute_error\"`.\n*Hint: vary the number of estimators between 1 and 60.*",
"_____no_output_____"
]
],
[
[
"# Write your code here.\nfrom sklearn.ensemble import AdaBoostRegressor\nfrom sklearn.model_selection import validation_curve\nimport pandas as pd\nimport numpy as np\n\nn_estimators = np.arange(1, 100, 2)\nn_estimators\n\nmodel = AdaBoostRegressor()\nmodel.get_params()\ntrain_scores, test_scores = validation_curve(\n model, data, target, param_name=\"n_estimators\", param_range=n_estimators,\n cv=5, scoring=\"neg_mean_absolute_error\", n_jobs=2)",
"_____no_output_____"
],
[
"train_errors, test_errors = -train_scores, -test_scores",
"_____no_output_____"
]
],
[
[
"Plot both the mean training and test errors. You can also plot the\nstandard deviation of the errors.\n*Hint: you can use `plt.errorbar`.*",
"_____no_output_____"
]
],
[
[
"# Write your code here.\nfrom matplotlib import pyplot as plt\n\nplt.errorbar(n_estimators, train_errors.mean(axis=1),\n yerr=train_errors.std(axis=1), label=\"Training error\")\n\nplt.errorbar(n_estimators, test_errors.mean(axis=1),\n yerr=train_errors.std(axis=1), label=\"Testing error\")",
"_____no_output_____"
]
],
[
[
"Plotting the validation curve, we can see that AdaBoost is not immune against\noverfitting. Indeed, there is an optimal number of estimators to be found.\nAdding too many estimators is detrimental for the statistical performance of\nthe model.",
"_____no_output_____"
],
[
"Repeat the experiment using a random forest instead of an AdaBoost regressor.",
"_____no_output_____"
]
],
[
[
"# Write your code here.\nfrom sklearn.ensemble import RandomForestRegressor\n\nmodel = RandomForestRegressor()\ntrain_scores, test_scores = validation_curve(\n model, data, target, param_name=\"n_estimators\", param_range=n_estimators,\n cv=5, scoring=\"neg_mean_absolute_error\", n_jobs=2)\n\n# PLOT\nplt.errorbar(n_estimators, train_errors.mean(axis=1),\n yerr=train_errors.std(axis=1), label=\"Training error\")\n\nplt.errorbar(n_estimators, test_errors.mean(axis=1),\n yerr=train_errors.std(axis=1), label=\"Testing error\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0b44faa53cc250ecd98b322b38afc345777bec9 | 495,490 | ipynb | Jupyter Notebook | README.ipynb | malleshi-9025/SmallPebble | 3b1eb340f39b0b8728668c2c6c8d1eadbcad4f10 | [
"Apache-2.0"
] | 84 | 2021-02-04T18:05:12.000Z | 2022-03-21T23:53:12.000Z | README.ipynb | malleshi-9025/SmallPebble | 3b1eb340f39b0b8728668c2c6c8d1eadbcad4f10 | [
"Apache-2.0"
] | 6 | 2021-03-27T06:06:02.000Z | 2022-03-23T17:43:13.000Z | README.ipynb | malleshi-9025/SmallPebble | 3b1eb340f39b0b8728668c2c6c8d1eadbcad4f10 | [
"Apache-2.0"
] | 10 | 2021-05-03T20:17:52.000Z | 2021-10-04T05:27:26.000Z | 342.662517 | 315,038 | 0.9204 | [
[
[
"# SmallPebble\n\n[](https://github.com/sradc/smallpebble/commits/) \n\n**Project status: unstable.**\n\n<br><p align=\"center\"><img src=\"https://raw.githubusercontent.com/sradc/SmallPebble/master/pebbles.jpg\"/></p><br>\n\nSmallPebble is a minimal automatic differentiation and deep learning library written from scratch in [Python](https://www.python.org/), using [NumPy](https://numpy.org/)/[CuPy](https://cupy.dev/).\n\nThe implementation is relatively small, and mainly in the file: [smallpebble.py](https://github.com/sradc/SmallPebble/blob/master/smallpebble/smallpebble.py). To help understand it, check out [this](https://sidsite.com/posts/autodiff/) introduction to autodiff, which presents an autodiff framework that works in the same way as SmallPebble (except using scalars instead of NumPy arrays).\n\nSmallPebble's *raison d'etre* is to be a simplified deep learning implementation,\nfor those who want to learn what’s under the hood of deep learning frameworks.\nHowever, because it is written in terms of vectorised NumPy/CuPy operations,\nit performs well enough for non-trivial models to be trained using it.\n\n**Highlights**\n- Relatively simple implementation.\n- Can run on GPU, using CuPy.\n- Various operations, such as matmul, conv2d, maxpool2d.\n- Array broadcasting support.\n- Eager or lazy execution.\n- Powerful API for creating models.\n- It's easy to add new SmallPebble functions.\n\n\n**Notes**\n\nGraphs are built implicitly via Python objects referencing Python objects.\nWhen `get_gradients` is called, autodiff is carried out on the whole sub-graph. The default array library is NumPy.\n\n---\n\n**Read on to see:**\n- Example models created and trained using SmallPebble.\n- A brief guide to using SmallPebble.\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nfrom tqdm.notebook import tqdm\nimport smallpebble as sp\nfrom smallpebble.misc import load_data",
"_____no_output_____"
]
],
[
[
"## Training a neural network to classify handwritten digits (MNIST)",
"_____no_output_____"
]
],
[
[
"\"Load the dataset, and create a validation set.\"\n\nX_train, y_train, _, _ = load_data('mnist') # load / download from openml.org\nX_train = X_train/255 # normalize\n\n# Seperate out data for validation.\nX = X_train[:50_000, ...]\ny = y_train[:50_000]\nX_eval = X_train[50_000:60_000, ...]\ny_eval = y_train[50_000:60_000]",
"_____no_output_____"
],
[
"\"Plot, to check we have the right data.\"\n\nplt.figure(figsize=(5,5))\nfor i in range(25):\n plt.subplot(5,5,i+1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(X_train[i,:].reshape(28,28), cmap='gray', vmin=0, vmax=1)\n\nplt.show()",
"_____no_output_____"
],
[
"\"Create a model, with two fully connected hidden layers.\"\n\nX_in = sp.Placeholder()\ny_true = sp.Placeholder()\n\nh = sp.linearlayer(28*28, 100)(X_in)\nh = sp.Lazy(sp.leaky_relu)(h)\nh = sp.linearlayer(100, 100)(h)\nh = sp.Lazy(sp.leaky_relu)(h)\nh = sp.linearlayer(100, 10)(h)\ny_pred = sp.Lazy(sp.softmax)(h)\nloss = sp.Lazy(sp.cross_entropy)(y_pred, y_true)\n\nlearnables = sp.get_learnables(y_pred)\n\nloss_vals = []\nvalidation_acc = []",
"_____no_output_____"
],
[
"\"Train model, while measuring performance on the validation dataset.\"\n\nNUM_ITERS = 300\nBATCH_SIZE = 200\n\neval_batch = sp.batch(X_eval, y_eval, BATCH_SIZE)\nadam = sp.Adam() # Adam optimization\n\nfor i, (xbatch, ybatch) in tqdm(enumerate(sp.batch(X, y, BATCH_SIZE)), total=NUM_ITERS):\n if i >= NUM_ITERS: break\n \n X_in.assign_value(sp.Variable(xbatch))\n y_true.assign_value(ybatch)\n \n loss_val = loss.run() # run the graph\n if np.isnan(loss_val.array):\n print(\"loss is nan, aborting.\")\n break\n loss_vals.append(loss_val.array)\n \n # Compute gradients, and use to carry out learning step:\n gradients = sp.get_gradients(loss_val)\n adam.training_step(learnables, gradients)\n \n # Compute validation accuracy:\n x_eval_batch, y_eval_batch = next(eval_batch)\n X_in.assign_value(sp.Variable(x_eval_batch))\n predictions = y_pred.run()\n predictions = np.argmax(predictions.array, axis=1)\n accuracy = (y_eval_batch == predictions).mean()\n validation_acc.append(accuracy)\n\n# Plot results:\nprint(f'Final validation accuracy: {np.mean(validation_acc[-10:])}')\nplt.figure(figsize=(14, 4))\nplt.subplot(1, 2, 1)\nplt.ylabel('Loss')\nplt.xlabel('Iteration')\nplt.plot(loss_vals)\nplt.subplot(1, 2, 2)\nplt.ylabel('Validation accuracy')\nplt.xlabel('Iteration')\nplt.suptitle('Neural network trained on MNIST, using SmallPebble.')\nplt.ylim([0, 1])\nplt.plot(validation_acc)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Training a convolutional neural network on CIFAR-10, using CuPy\nThis was run on [Google Colab](https://colab.research.google.com/), with a GPU.",
"_____no_output_____"
]
],
[
[
"\"Load the CIFAR dataset.\"\n\nX_train, y_train, _, _ = load_data('cifar') # load/download from openml.org\nX_train = X_train/255 # normalize",
"_____no_output_____"
],
[
"\"\"\"Plot, to check it's the right data.\n\n(This cell's code is from: https://www.tensorflow.org/tutorials/images/cnn#verify_the_data)\n\"\"\"\n\nclass_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',\n 'dog', 'frog', 'horse', 'ship', 'truck']\n\nplt.figure(figsize=(8,8))\nfor i in range(25):\n plt.subplot(5,5,i+1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(X_train[i,:].reshape(32,32,3))\n plt.xlabel(class_names[y_train[i]])\n\nplt.show()",
"_____no_output_____"
],
[
"\"Switch array library to CuPy, so can use GPU.\"\n\nimport cupy\n\nsp.use(cupy)\n\nprint(sp.array_library.library.__name__) # should be 'cupy'",
"\ncupy\n"
],
[
"\"Convert data to CuPy arrays\"\n\nX_train = cupy.array(X_train)\ny_train = cupy.array(y_train)\n\n# Seperate out data for validation as before.\nX = X_train[:45_000, ...]\ny = y_train[:45_000]\nX_eval = X_train[45_000:50_000, ...]\ny_eval = y_train[45_000:50_000]",
"_____no_output_____"
],
[
"\"\"\"Define a model.\"\"\"\n\nX_in = sp.Placeholder()\ny_true = sp.Placeholder()\n\nh = sp.convlayer(height=3, width=3, depth=3, n_kernels=32)(X_in)\nh = sp.Lazy(sp.leaky_relu)(h)\nh = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)\n\nh = sp.convlayer(3, 3, 32, 128, padding='VALID')(h)\nh = sp.Lazy(sp.leaky_relu)(h)\nh = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)\n\nh = sp.convlayer(3, 3, 128, 128, padding='VALID')(h)\nh = sp.Lazy(sp.leaky_relu)(h)\nh = sp.Lazy(lambda a: sp.maxpool2d(a, 2, 2, strides=[2, 2]))(h)\n\nh = sp.Lazy(lambda x: sp.reshape(x, [-1, 3*3*128]))(h)\nh = sp.linearlayer(3*3*128, 10)(h)\nh = sp.Lazy(sp.softmax)(h)\n\ny_pred = h\nloss = sp.Lazy(sp.cross_entropy)(y_pred, y_true)\n\nlearnables = sp.get_learnables(y_pred)\n\nloss_vals = []\nvalidation_acc = []\n\n# Check we get the expected dimensions\nX_in.assign_value(sp.Variable(X[0:3, :].reshape([-1, 32, 32, 3])))\nh.run().shape",
"_____no_output_____"
]
],
[
[
"Train the model.",
"_____no_output_____"
]
],
[
[
"NUM_ITERS = 3000\nBATCH_SIZE = 128\n\neval_batch = sp.batch(X_eval, y_eval, BATCH_SIZE)\nadam = sp.Adam()\n\nfor i, (xbatch, ybatch) in tqdm(enumerate(sp.batch(X, y, BATCH_SIZE)), total=NUM_ITERS):\n if i >= NUM_ITERS: break\n \n xbatch_images = xbatch.reshape([-1, 32, 32, 3])\n X_in.assign_value(sp.Variable(xbatch_images))\n y_true.assign_value(ybatch)\n \n loss_val = loss.run()\n if np.isnan(loss_val.array):\n print(\"Aborting, loss is nan.\")\n break\n loss_vals.append(loss_val.array)\n \n # Compute gradients, and carry out learning step.\n gradients = sp.get_gradients(loss_val) \n adam.training_step(learnables, gradients)\n \n # Compute validation accuracy:\n x_eval_batch, y_eval_batch = next(eval_batch)\n X_in.assign_value(sp.Variable(x_eval_batch.reshape([-1, 32, 32, 3])))\n predictions = y_pred.run()\n predictions = np.argmax(predictions.array, axis=1)\n accuracy = (y_eval_batch == predictions).mean()\n validation_acc.append(accuracy)\n\nprint(f'Final validation accuracy: {np.mean(validation_acc[-10:])}')\nplt.figure(figsize=(14, 4))\nplt.subplot(1, 2, 1)\nplt.ylabel('Loss')\nplt.xlabel('Iteration')\nplt.plot(loss_vals)\nplt.subplot(1, 2, 2)\nplt.ylabel('Validation accuracy')\nplt.xlabel('Iteration')\nplt.suptitle('CNN trained on CIFAR-10, using SmallPebble.')\nplt.ylim([0, 1])\nplt.plot(validation_acc)\nplt.show()",
"_____no_output_____"
]
],
[
[
"It looks like we could improve our results by training for longer (and we could improve our model architecture).",
"_____no_output_____"
],
[
"---\n\n# Brief guide to using SmallPebble\n\nSmallPebble provides the following building blocks to make models with:\n\n- `sp.Variable`\n- Operations, such as `sp.add`, `sp.mul`, etc.\n- `sp.get_gradients`\n- `sp.Lazy`\n- `sp.Placeholder` (this is really just `sp.Lazy` on the identity function)\n- `sp.learnable`\n- `sp.get_learnables`\n\nThe following examples show how these are used.\n",
"_____no_output_____"
],
[
"## Switching between NumPy and CuPy\n\nWe can dynamically switch between NumPy and CuPy. (Assuming you have a CuPy compatible GPU and CuPy set up. Note, CuPy is available on Google Colab, if you change the runtime to GPU.)",
"_____no_output_____"
]
],
[
[
"import cupy\nimport numpy\nimport smallpebble as sp\n \n# Switch to CuPy\nsp.use(cupy)\nprint(sp.array_library.library.__name__) # should be 'cupy'\n\n# Switch back to NumPy:\nsp.use(numpy)\nprint(sp.array_library.library.__name__) # should be 'numpy'",
"cupy\nnumpy\n"
]
],
[
[
"## sp.Variable & sp.get_gradients \n\nWith SmallPebble, you can:\n\n- Wrap NumPy arrays in `sp.Variable`\n- Apply SmallPebble operations (e.g. `sp.matmul`, `sp.add`, etc.)\n- Compute gradients with `sp.get_gradients`",
"_____no_output_____"
]
],
[
[
"a = sp.Variable(np.random.random([2, 2]))\nb = sp.Variable(np.random.random([2, 2]))\nc = sp.Variable(np.random.random([2]))\ny = sp.mul(a, b) + c\nprint('y.array:\\n', y.array)\n\ngradients = sp.get_gradients(y)\ngrad_a = gradients[a]\ngrad_b = gradients[b]\ngrad_c = gradients[c]\nprint('grad_a:\\n', grad_a)\nprint('grad_b:\\n', grad_b)\nprint('grad_c:\\n', grad_c)",
"y.array:\n [[1.32697776 1.24689392]\n [1.25317932 1.05037433]]\ngrad_a:\n [[0.50232192 0.99209074]\n [0.42936606 0.19027664]]\ngrad_b:\n [[0.95442445 0.34679685]\n [0.94471809 0.7753676 ]]\ngrad_c:\n [2. 2.]\n"
]
],
[
[
"Note that `y` is computed straight away, i.e. the (forward) computation happens immediately.\n\nAlso note that `y` is a sp.Variable and we could continue to carry out SmallPebble operations on it.",
"_____no_output_____"
],
[
"## sp.Lazy & sp.Placeholder\n\nLazy graphs are constructed using `sp.Lazy` and `sp.Placeholder`. ",
"_____no_output_____"
]
],
[
[
"lazy_node = sp.Lazy(lambda a, b: a + b)(1, 2)\nprint(lazy_node)\nprint(lazy_node.run())",
"<smallpebble.smallpebble.Lazy object at 0x7f15db527550>\n3\n"
],
[
"a = sp.Lazy(lambda a: a)(2)\ny = sp.Lazy(lambda a, b, c: a * b + c)(a, 3, 4)\nprint(y)\nprint(y.run())",
"<smallpebble.smallpebble.Lazy object at 0x7f15db26ea50>\n10\n"
]
],
[
[
"Forward computation does not happen immediately - only when .run() is called.",
"_____no_output_____"
]
],
[
[
"a = sp.Placeholder()\nb = sp.Variable(np.random.random([2, 2]))\ny = sp.Lazy(sp.matmul)(a, b)\n\na.assign_value(sp.Variable(np.array([[1,2], [3,4]])))\n\nresult = y.run()\nprint('result.array:\\n', result.array)",
"result.array:\n [[1.96367495 2.26668698]\n [3.94895132 5.3053362 ]]\n"
]
],
[
[
"You can use .run() as many times as you like. \n\nLet's change the placeholder value and re-run the graph:",
"_____no_output_____"
]
],
[
[
"a.assign_value(sp.Variable(np.array([[10,20], [30,40]])))\nresult = y.run()\nprint('result.array:\\n', result.array)",
"result.array:\n [[19.63674952 22.6668698 ]\n [39.48951324 53.053362 ]]\n"
]
],
[
[
"Finally, let's compute gradients:",
"_____no_output_____"
]
],
[
[
"gradients = sp.get_gradients(result)",
"_____no_output_____"
]
],
[
[
"Note that `sp.get_gradients` is called on `result`, \nwhich is a `sp.Variable`, \nnot on `y`, which is a `sp.Lazy` instance.",
"_____no_output_____"
],
[
"## sp.learnable & sp.get_learnables\nUse `sp.learnable` to flag parameters as learnable, \nallowing them to be extracted from a lazy graph with `sp.get_learnables`.\n\nThis enables a workflow of: building a model, while flagging parameters as learnable, and then extracting all the parameters in one go at the end.\n",
"_____no_output_____"
]
],
[
[
"a = sp.Placeholder()\nb = sp.learnable(sp.Variable(np.random.random([2, 1])))\ny = sp.Lazy(sp.matmul)(a, b)\ny = sp.Lazy(sp.add)(y, sp.learnable(sp.Variable(np.array([5]))))\n\nlearnables = sp.get_learnables(y)\n\nfor learnable in learnables:\n print(learnable)",
"<smallpebble.smallpebble.Variable object at 0x7f157a263b10>\n<smallpebble.smallpebble.Variable object at 0x7f15d2a4ccd0>\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0b484ae702875dc57098f8e188b7dd7f9a8ffe3 | 4,109 | ipynb | Jupyter Notebook | 41-warmup-blank_introduction_to_machine_learning.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
] | 6 | 2020-04-13T19:22:18.000Z | 2021-04-20T18:20:13.000Z | 41-warmup-blank_introduction_to_machine_learning.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
] | null | null | null | 41-warmup-blank_introduction_to_machine_learning.ipynb | hanisaf/advanced-data-management-and-analytics | e7bffda5cad91374a14df1a65f95e6a25f72cc41 | [
"MIT"
] | 10 | 2020-05-12T01:02:32.000Z | 2022-02-28T17:04:37.000Z | 24.753012 | 97 | 0.394257 | [
[
[
"## In this exercise we will try to re-create the decision tree from data\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sklearn import tree\nfrom sklearn.tree import export_text",
"_____no_output_____"
],
[
"data = pd.read_csv(\"data/PlayTennis.csv\")",
"_____no_output_____"
]
],
[
[
"Train a decision tree using the data and then print the tree\n\n*Note* You will need to change the values from string to numbers before training the tree\n",
"_____no_output_____"
]
],
[
[
"data.head()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b48914e5c6fabba3c8f72f1830b7407f4e6ddf | 523,612 | ipynb | Jupyter Notebook | notebooks/integrate_seasonal_et_p.ipynb | ecohydro/rhone-ecostress | fa72f4c2716f40a860551ef4073fbaa5b004c7f5 | [
"MIT"
] | 2 | 2020-10-12T21:46:17.000Z | 2022-02-12T03:51:10.000Z | notebooks/integrate_seasonal_et_p.ipynb | ecohydro/rhone-ecostress | fa72f4c2716f40a860551ef4073fbaa5b004c7f5 | [
"MIT"
] | null | null | null | notebooks/integrate_seasonal_et_p.ipynb | ecohydro/rhone-ecostress | fa72f4c2716f40a860551ef4073fbaa5b004c7f5 | [
"MIT"
] | 2 | 2020-07-31T19:35:54.000Z | 2022-02-04T13:25:24.000Z | 96.287606 | 185,276 | 0.67012 | [
[
[
"import rioxarray as rio\nimport xarray as xr\nimport glob\nimport os\nimport numpy as np\nimport requests\nimport geopandas as gpd\nfrom pathlib import Path\nfrom datetime import datetime\nfrom rasterio.enums import Resampling\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nsite = \"BRC\"\n\n# Change site name\nchirps_seas_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_season_precip_data_brc')\neeflux_seas_int_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_growing_season_integrated_brc') \nchirps_wy_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/wy_total_chirps_brc')\neeflux_seas_mean_out_dir = Path('/home/serdp/rhone/rhone-ecostress/rasters/ee_season_mean_brc') \n\nall_scenes_f_precip = Path('/scratch/waves/rhone-ecostress/rasters/chirps-clipped')\nall_scenes_f_et = Path('/home/serdp/rhone/rhone-ecostress/rasters/eeflux/BRC') # Change file path based on site\n\nall_precip_paths = list(all_scenes_f_precip.glob(\"*\"))\nall_et_paths = list(all_scenes_f_et.glob(\"*.tif\")) # Variable name agnostic to site?",
"_____no_output_____"
],
[
"# for some reason the fll value is not correct. this is the correct bad value to mask by\ntestf = all_precip_paths[0]\nx = rio.open_rasterio(testf)\nbadvalue = np.unique(x.where(x != x._FillValue).sel(band=1))[0]\n\ndef chirps_path_date(path):\n _, _, year, month, day, _ = path.name.split(\".\") \n day = day.split(\"-\")[0]\n return datetime(int(year), int(month), int(day))\n\n\ndef open_chirps(path):\n data_array = rio.open_rasterio(path) #chunks makes i lazyily executed\n data_array = data_array.sel(band=1).drop(\"band\") # gets rid of old coordinate dimension since we need bands to have unique coord ids\n data_array[\"date\"] = chirps_path_date(path) # makes a new coordinate\n return data_array.expand_dims({\"date\":1}) # makes this coordinate a dimension\n\n\n\n### data is not tiled so not a good idea to use chunking\n#https://github.com/pydata/xarray/issues/2314\n\nimport rasterio\nwith rasterio.open(testf) as src:\n print(src.profile)\n\nlen(all_precip_paths) * 41.7 / 10e3 # convert from in to mm\n\n%timeit open_chirps(testf)\n\nall_daily_precip_path = \"/home/serdp/ravery/rhone-ecostress/netcdfs/all_chirps_daily_i.nc\"\n\nif Path(all_daily_precip_path).exists():\n \n all_chirps_arr = xr.open_dataarray(all_daily_precip_path)\n all_chirps_arr = all_chirps_arr.sortby(\"date\")\nelse:\n\n daily_chirps_arrs = []\n\n for path in all_precip_paths:\n\n daily_chirps_arrs.append(open_chirps(path)) \n \n all_chirps_arr = xr.concat(daily_chirps_arrs, dim=\"date\")\n \n all_chirps_arr = all_chirps_arr.sortby(\"date\")\n\n all_chirps_arr.to_netcdf(all_daily_precip_path)\n\ndef eeflux_path_date(path):\n year, month, day, _, _ = path.name.split(\"-\") # Change this line accordingly based on format of eeflux dates\n return datetime(int(year), int(month), int(day))\n\ndef open_eeflux(path, da_for_match):\n data_array = rio.open_rasterio(path) #chunks makes i lazyily executed\n data_array.rio.reproject_match(da_for_match)\n data_array = data_array.sel(band=1).drop(\"band\") # gets rid of old coordinate dimension since we need bands to have unique coord ids\n data_array[\"date\"] = eeflux_path_date(path) # makes a new coordinate\n return data_array.expand_dims({\"date\":1}) # makes this coordinate a dimension\n\n# The following lines seem to write the lists of rasters to netcdf files? Do we need to replicate for chirps?\nda_for_match = rio.open_rasterio(all_et_paths[0])\ndaily_eeflux_arrs = [open_eeflux(path, da_for_match) for path in all_et_paths]\nall_eeflux_arr = xr.concat(daily_eeflux_arrs, dim=\"date\")\nall_daily_eeflux_path = \"/home/serdp/ravery/rhone-ecostress/netcdfs/all_eeflux_daily_i.nc\"\nall_eeflux_arr.to_netcdf(all_daily_eeflux_path)\n\nall_eeflux_arr[-3,:,:].plot.imshow()\n\nall_eeflux_arr = all_eeflux_arr.sortby(\"date\")",
"{'driver': 'GTiff', 'dtype': 'float32', 'nodata': -3.4e+38, 'width': 55, 'height': 89, 'count': 1, 'crs': CRS.from_wkt('PROJCS[\"unknown\",GEOGCS[\"unknown\",DATUM[\"Unknown_based_on_GRS80_ellipsoid\",SPHEROID[\"GRS 1980\",6378137,298.257222101004,AUTHORITY[\"EPSG\",\"7019\"]],TOWGS84[0,0,0,0,0,0,0]],PRIMEM[\"Greenwich\",0],UNIT[\"degree\",0.0174532925199433,AUTHORITY[\"EPSG\",\"9122\"]]],PROJECTION[\"Lambert_Conformal_Conic_2SP\"],PARAMETER[\"latitude_of_origin\",46.5],PARAMETER[\"central_meridian\",3],PARAMETER[\"standard_parallel_1\",49],PARAMETER[\"standard_parallel_2\",44],PARAMETER[\"false_easting\",700000],PARAMETER[\"false_northing\",6600000],UNIT[\"metre\",1,AUTHORITY[\"EPSG\",\"9001\"]],AXIS[\"Easting\",EAST],AXIS[\"Northing\",NORTH]]'), 'transform': Affine(3940.0, 0.0, 752344.6729321405,\n 0.0, -5550.0, 6682060.744602234), 'tiled': False, 'compress': 'lzw', 'interleave': 'band'}\n79.7 ms ± 374 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)\n"
],
[
"ey = max(all_eeflux_arr['date.year'].values)\ney\nsy = min(all_eeflux_arr['date.year'].values)\nsy",
"_____no_output_____"
],
[
"all_eeflux_arr['date.dayofyear'].values",
"_____no_output_____"
],
[
"# THIS IS IMPORTANT\ndef years_list(all_arr):\n ey = max(all_arr['date.year'].values)\n sy = min(all_arr['date.year'].values)\n start_years = range(sy, ey)\n end_years = range(sy+1, ey+1) # Change to sy+1, ey+1 for across-calendar-year (e.g. winter) calculations\n return list(zip(start_years, end_years))\n\ndef group_by_custom_doy(all_arr, doy_start, doy_end):\n start_end_years = years_list(all_arr)\n water_year_arrs = []\n for water_year in start_end_years:\n start_mask = ((all_arr['date.dayofyear'].values > doy_start) & (all_arr['date.year'].values == water_year[0]))\n end_mask = ((all_arr['date.dayofyear'].values < doy_end) & (all_arr['date.year'].values == water_year[1]))\n water_year_arrs.append(all_arr[start_mask | end_mask]) # | = or, & = and\n return water_year_arrs\n\ndef group_by_season(all_arr, doy_start, doy_end):\n yrs = np.unique(all_arr['date.year'])\n season_arrs = []\n for yr in yrs:\n start_mask = ((all_arr['date.dayofyear'].values >= doy_start) & (all_arr['date.year'].values == yr))\n end_mask = ((all_arr['date.dayofyear'].values <= doy_end) & (all_arr['date.year'].values == yr))\n season_arrs.append(all_arr[start_mask & end_mask])\n return season_arrs",
"_____no_output_____"
],
[
"# THIS IS IMPORTANT\ndoystart = 125 # Edit these variables to change doy length of year\ndoyend = 275\n# eeflux_water_year_arrs = group_by_custom_doy(all_eeflux_arr, doystart, doyend) # Replaced by eeflux_seas_arrs below\nchirps_water_year_arrs = group_by_custom_doy(all_chirps_arr, doyend, doystart)",
"_____no_output_____"
],
[
"eeflux_seas_arrs = group_by_season(all_eeflux_arr, doystart, doyend)\neeflux_seas_arrs",
"_____no_output_____"
],
[
"chirps_water_year_arrs[-1]",
"_____no_output_____"
],
[
"fig = plt.figure()\nplt.plot(wy_list,[arr.mean() for arr in chirps_wy_sums],'.')\nplt.ylabel('WY Precipitation (mm)')",
"_____no_output_____"
],
[
"# Creates figure of ET availability\ngroup_counts = list(map(lambda x: len(x['date']), water_year_arrs))\nyear_tuples = years_list(all_eeflux_arr)\n\nindexes = np.arange(len(year_tuples))\nplt.bar(indexes, group_counts)\ndegrees = 80\nplt.xticks(indexes, year_tuples, rotation=degrees, ha=\"center\")\nplt.title(\"Availability of EEFLUX between DOY 125 and 275\")\nplt.savefig(\"eeflux_availability.png\")\n\n# Figure below shows empty years in 85, 88, 92, 93, 96; no winter precip rasters generated for these years b/c no ET data w/in winter window",
"_____no_output_____"
],
[
"def sum_seasonal_precip(precip_arr, eeflux_group_arr):\n return precip_arr.sel(date=slice(eeflux_group_arr.date.min(), eeflux_group_arr.date.max())).sum(dim=\"date\")\n# This is matching up precip w/ available ET window for each year",
"_____no_output_____"
],
[
"for index, eeflux_group in enumerate(eeflux_seas_arrs):\n if len(eeflux_group['date']) > 0:\n seasonal_precip = sum_seasonal_precip(all_chirps_arr, eeflux_group) # Variable/array name matters here\n seasonal_et = eeflux_group.integrate(coord=\"date\", datetime_unit=\"D\")\n year = eeflux_group['date.year'].values[0]\n et_doystart = eeflux_group['date.dayofyear'].values[0]\n et_doyend = eeflux_group['date.dayofyear'].values[-1]\n pname = os.path.join(chirps_seas_out_dir,f\"seas_chirps_{site}_{year}_{et_doystart}_{et_doyend}.tif\") #Edit output raster labels\n eename = os.path.join(eeflux_seas_int_out_dir, f\"seasonal_eeflux_integrated_{site}_{year}_{et_doystart}_{et_doyend}.tif\")\n seasonal_precip.rio.to_raster(pname)\n seasonal_et.rio.to_raster(eename)\n# This chunk actually outputs the rasters",
"_____no_output_____"
],
[
"## Elmera Additions for winter precip:\nfor index, (eeflux_group,chirps_group) in enumerate(zip(eeflux_seas_arrs,chirps_water_year_arrs[3:])): #changed eeflux_group to eeflux_seas_arrs & changed from water_year_arrs to season_arrs\n if len(eeflux_group['date']) > 0: # eeflux_group to eeflux_seas_arrs\n mean_seas_et = eeflux_group.mean(dim='date',skipna=False)\n chirps_wy_sum = chirps_group.sum(dim='date',skipna=False)\n # seasonal_precip = sum_seasonal_precip(chirps_water_year_arrs, eeflux_seas_arr) # Here's where above fxn is applied to rasters, need to replace eeflux_group\n year = eeflux_group['date.year'].values[0] \n pname = os.path.join(chirps_wy_out_dir,f\"wy_total_chirps_{site}_{year}.tif\") #Edit output raster labels\n eename = os.path.join(eeflux_seas_mean_out_dir,f\"mean_daily_seas_et_{site}_{year}.tif\")\n chirps_wy_sum.rio.to_raster(pname)\n mean_seas_et.rio.to_raster(eename)\n# This chunk actually outputs the rasters, ET lines removed - including seasonal_precip line?",
"_____no_output_____"
],
[
"[arr['date.year'] for arr in chirps_water_year_arrs]",
"_____no_output_____"
],
[
"seasonal_precip # This just shows the array - corner cells have empty values b/c of projection mismatch @ edge of raster",
"_____no_output_____"
],
[
"water_year_arrs[0][0].plot.imshow()",
"_____no_output_____"
],
[
"water_year_arrs[0].integrate(dim=\"date\", datetime_unit=\"D\").plot.imshow()\n# This chunk does the actual integration",
"<ipython-input-18-2edd85b98b08>:1: FutureWarning: The `dim` keyword argument to `DataArray.integrate` is being replaced with `coord`, for consistency with `Dataset.integrate`. Please pass `coord` instead. `dim` will be removed in version 0.19.0.\n water_year_arrs[0].integrate(dim=\"date\", datetime_unit=\"D\").plot.imshow()\n"
],
[
"all_eeflux_arr.integrate(dim=\"date\", datetime_unit=\"D\")",
"<ipython-input-19-1dea8731bb8e>:1: FutureWarning: The `dim` keyword argument to `DataArray.integrate` is being replaced with `coord`, for consistency with `Dataset.integrate`. Please pass `coord` instead. `dim` will be removed in version 0.19.0.\n all_eeflux_arr.integrate(dim=\"date\", datetime_unit=\"D\")\n"
],
[
"import pandas as pd\nimport numpy as np\n\nlabels = ['<=2', '3-9', '>=10']\nbins = [0,2,9, np.inf]\npd.cut(all_eeflux_arr, bins, labels=labels)",
"_____no_output_____"
],
[
"all_eeflux_arr",
"_____no_output_____"
],
[
"import pandas as pd\n\nall_scene_ids = [str(i) for i in list(all_scenes_f.glob(\"L*\"))]\ndf = pd.DataFrame({\"scene_id\":all_scene_ids}).reindex()\nsplit_vals_series = df.scene_id.str.split(\"/\")\n\ndff = pd.DataFrame(split_vals_series.to_list(), columns=['_', '__', '___', '____', '_____', '______', 'fname'])\n\ndf['date'] = dff['fname'].str.slice(10,18)\n\ndf['pathrow'] = dff['fname'].str.slice(4,10)\n\ndf['sensor'] = dff['fname'].str.slice(0,4)\n\ndf['datetime'] = pd.to_datetime(df['date'])\n\ndf = df.set_index(\"datetime\").sort_index()",
"_____no_output_____"
],
[
"marc_df = df['2014-01-01':'2019-12-31']",
"_____no_output_____"
],
[
"marc_df = marc_df[marc_df['sensor']==\"LC08\"]",
"_____no_output_____"
],
[
"x.where(x != badvalue).sel(band=1).plot.imshow()",
"_____no_output_____"
],
[
"# Evan additions\n\nyear_tuples = years_list(all_eeflux_arr)\nyear_tuples\n",
"_____no_output_____"
],
[
"# Winter precip calculations\nyear_tuples_p = years_list(all_chirps_arr)\nyear_tuples_p",
"_____no_output_____"
],
[
"def group_p_by_custom_doy(all_chirps_arr, doy_start, doy_end):\n start_end_years = years_list(all_chirps_arr)\n water_year_arrs = []\n for water_year in start_end_years:\n start_mask = ((all_chirps_arr['date.dayofyear'].values > doy_start) & (all_chirps_arr['date.year'].values == water_year[0]))\n end_mask = ((all_chirps_arr['date.dayofyear'].values < doy_end) & (all_chirps_arr['date.year'].values == water_year[0]))\n water_year_arrs.append(all_chirps_arr[start_mask | end_mask])\n return water_year_arrs\ndoystart = 275 # Edit these variables to change doy length of year\ndoyend = 125\nwater_year_arrs = group_p_by_custom_doy(all_chirps_arr, doystart, doyend)\nwater_year_arrs",
"_____no_output_____"
],
[
"def sum_seasonal_precip(precip_arr, eeflux_group_arr):\n return precip_arr.sel(date=slice(eeflux_group_arr.date.min(), eeflux_group_arr.date.max())).sum(dim=\"date\")\n# This is matching up precip w/ available ET window for each year, need to figure out what to feed in for 2nd variable",
"_____no_output_____"
],
[
"for index, eeflux_group in enumerate(water_year_arrs):\n if len(eeflux_group['date']) > 0:\n seasonal_precip = sum_seasonal_precip(all_chirps_arr, eeflux_group) # Here's where above fxn is applied to rasters, need to replace eeflux_group\n year_range = year_tuples_p[index]\n pname = f\"winter_chirps_{year_range[0]}_{year_range[1]}_{doystart}_{doyend}.tif\" #Edit output raster labels\n seasonal_precip.rio.to_raster(pname)\n# This chunk actually outputs the rasters, ET lines removed",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b48b062f7734c200cd9f930ee405ddfe12f145 | 18,232 | ipynb | Jupyter Notebook | database/tasks/How to create a QQ-plot/Python, using SciPy.ipynb | nathancarter/how2data | 7d4f2838661f7ce98deb1b8081470cec5671b03a | [
"MIT"
] | null | null | null | database/tasks/How to create a QQ-plot/Python, using SciPy.ipynb | nathancarter/how2data | 7d4f2838661f7ce98deb1b8081470cec5671b03a | [
"MIT"
] | null | null | null | database/tasks/How to create a QQ-plot/Python, using SciPy.ipynb | nathancarter/how2data | 7d4f2838661f7ce98deb1b8081470cec5671b03a | [
"MIT"
] | 2 | 2021-07-18T19:01:29.000Z | 2022-03-29T06:47:11.000Z | 161.345133 | 15,390 | 0.908896 | [
[
[
"---\nauthor:\n - Elizabeth Czarniak ([email protected])\n - Nathan Carter ([email protected])\n---",
"_____no_output_____"
],
[
"We're going to use some fake data here by generating random numbers, but you can replace our fake data with your real data in the code below.",
"_____no_output_____"
]
],
[
[
"# Replace this with your data, such as a variable or column in a DataFrame\nimport numpy as np\nvalues = np.random.normal(0, 1, 50) # 50 random values",
"_____no_output_____"
]
],
[
[
"If the data is normally distributed, then we expect that the QQ plot will show the observed values (blue dots) falling very clsoe to the red line (the quantiles for the normal distribution).",
"_____no_output_____"
]
],
[
[
"from scipy import stats\nimport matplotlib.pyplot as plt\n\nstats.probplot(values, dist=\"norm\", plot=plt)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Our observed values fall pretty close to the reference line. In this case, we expected that, because we created fake data that was normally distributed. But for real data, it may not stay so close to the red line.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b48e50d28bd66d44de17e22da1479415d82b73 | 5,820 | ipynb | Jupyter Notebook | qc-training-notebooks/index.ipynb | sanger-pathogens/QC-training | 657e1e6ccb40d1d91842ee21bb37940c5d1a395c | [
"CC-BY-4.0"
] | 1 | 2021-11-07T21:56:21.000Z | 2021-11-07T21:56:21.000Z | qc-training-notebooks/index.ipynb | sanger-pathogens/QC-training | 657e1e6ccb40d1d91842ee21bb37940c5d1a395c | [
"CC-BY-4.0"
] | 2 | 2021-01-11T12:27:10.000Z | 2021-01-13T09:34:58.000Z | qc-training-notebooks/index.ipynb | sanger-pathogens/QC-training | 657e1e6ccb40d1d91842ee21bb37940c5d1a395c | [
"CC-BY-4.0"
] | null | null | null | 35.060241 | 555 | 0.639863 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b4a29f1f26d9083bf69ba6b415b81b0a8174d3 | 22,106 | ipynb | Jupyter Notebook | 6_dqn_cartpole.ipynb | yuhonghong66/chainer-notebooks | 639db107034346055b51af98fafdffc9f4bd52d2 | [
"MIT"
] | 20 | 2017-10-08T12:02:59.000Z | 2021-01-13T17:57:33.000Z | 6_dqn_cartpole.ipynb | mitmul/DSVM-Chainer-Notebooks | 639db107034346055b51af98fafdffc9f4bd52d2 | [
"MIT"
] | null | null | null | 6_dqn_cartpole.ipynb | mitmul/DSVM-Chainer-Notebooks | 639db107034346055b51af98fafdffc9f4bd52d2 | [
"MIT"
] | 6 | 2017-10-09T15:36:48.000Z | 2020-11-29T21:56:31.000Z | 41.552632 | 614 | 0.617796 | [
[
[
"# ChainerRL Quickstart Guide\n\nThis is a quickstart guide for users who just want to try ChainerRL for the first time.\n\nIf you have not yet installed ChainerRL, run the command below to install it:",
"_____no_output_____"
]
],
[
[
"%%bash\npip install chainerrl",
"Collecting chainerrl\n Downloading chainerrl-0.2.0.tar.gz (56kB)\nCollecting cached-property (from chainerrl)\n Downloading cached_property-1.3.1-py2.py3-none-any.whl\nRequirement already satisfied: chainer>=2.0.0 in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainerrl)\nCollecting future (from chainerrl)\n Downloading future-0.16.0.tar.gz (824kB)\nCollecting gym>=0.7.3 (from chainerrl)\n Downloading gym-0.9.3.tar.gz (157kB)\nRequirement already satisfied: numpy>=1.10.4 in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainerrl)\nRequirement already satisfied: pillow in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainerrl)\nRequirement already satisfied: scipy in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainerrl)\nRequirement already satisfied: six>=1.9.0 in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainer>=2.0.0->chainerrl)\nRequirement already satisfied: protobuf>=2.6.0 in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainer>=2.0.0->chainerrl)\nRequirement already satisfied: mock in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainer>=2.0.0->chainerrl)\nRequirement already satisfied: nose in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainer>=2.0.0->chainerrl)\nRequirement already satisfied: filelock in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from chainer>=2.0.0->chainerrl)\nRequirement already satisfied: requests>=2.0 in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from gym>=0.7.3->chainerrl)\nCollecting pyglet>=1.2.0 (from gym>=0.7.3->chainerrl)\n Downloading pyglet-1.2.4-py3-none-any.whl (964kB)\nRequirement already satisfied: olefile in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from pillow->chainerrl)\nRequirement already satisfied: setuptools in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages/setuptools-27.2.0-py3.6.egg (from protobuf>=2.6.0->chainer>=2.0.0->chainerrl)\nRequirement already satisfied: pbr>=0.11 in /home/shunta/.pyenv/versions/anaconda3-4.4.0/lib/python3.6/site-packages (from mock->chainer>=2.0.0->chainerrl)\nBuilding wheels for collected packages: chainerrl, future, gym\n Running setup.py bdist_wheel for chainerrl: started\n Running setup.py bdist_wheel for chainerrl: finished with status 'done'\n Stored in directory: /home/shunta/.cache/pip/wheels/50/e1/16/d6879538da7fe0053f5b61c3d1f4e1b009464d3564b99c792c\n Running setup.py bdist_wheel for future: started\n Running setup.py bdist_wheel for future: finished with status 'done'\n Stored in directory: /home/shunta/.cache/pip/wheels/c2/50/7c/0d83b4baac4f63ff7a765bd16390d2ab43c93587fac9d6017a\n Running setup.py bdist_wheel for gym: started\n Running setup.py bdist_wheel for gym: finished with status 'done'\n Stored in directory: /home/shunta/.cache/pip/wheels/2b/16/05/14202d3528fb14912254fe7062bfc8b061ade8de9409f1abd0\nSuccessfully built chainerrl future gym\nInstalling collected packages: cached-property, future, pyglet, gym, chainerrl\nSuccessfully installed cached-property-1.3.1 chainerrl-0.2.0 future-0.16.0 gym-0.9.3 pyglet-1.2.4\n"
]
],
[
[
"If you have already installed ChainerRL, let's begin!\n\nFirst, you need to import necessary modules. The module name of ChainerRL is `chainerrl`. Let's import `gym` and `numpy` as well since they are used later.",
"_____no_output_____"
]
],
[
[
"import chainer\nimport chainer.functions as F\nimport chainer.links as L\nimport chainerrl\nimport gym\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"ChainerRL can be used for any problems if they are modeled as \"environments\". [OpenAI Gym](https://github.com/openai/gym) provides various kinds of benchmark environments and defines the common interface among them. ChainerRL uses a subset of the interface. Specifically, an environment must define its observation space and action space and have at least two methods: `reset` and `step`.\n\n- `env.reset` will reset the environment to the initial state and return the initial observation.\n- `env.step` will execute a given action, move to the next state and return four values:\n - a next observation\n - a scalar reward\n - a boolean value indicating whether the current state is terminal or not\n - additional information\n- `env.render` will render the current state.\n\nLet's try 'CartPole-v0', which is a classic control problem. You can see below that its observation space consists of four real numbers while its action space consists of two discrete actions.",
"_____no_output_____"
]
],
[
[
"env = gym.make('CartPole-v0')\nprint('observation space:', env.observation_space)\nprint('action space:', env.action_space)\n\nobs = env.reset()\nenv.render(close=True)\nprint('initial observation:', obs)\n\naction = env.action_space.sample()\nobs, r, done, info = env.step(action)\nprint('next observation:', obs)\nprint('reward:', r)\nprint('done:', done)\nprint('info:', info)",
"[2017-09-23 05:17:39,776] Making new env: CartPole-v0\n"
]
],
[
[
"Now you have defined your environment. Next, you need to define an agent, which will learn through interactions with the environment.\n\nChainerRL provides various agents, each of which implements a deep reinforcement learning algorithm.\n\nTo use [DQN (Deep Q-Network)](http://dx.doi.org/10.1038/nature14236), you need to define a Q-function that receives an observation and returns an expected future return for each action the agent can take. In ChainerRL, you can define your Q-function as `chainer.Link` as below. Note that the outputs are wrapped by `chainerrl.action_value.DiscreteActionValue`, which implements `chainerrl.action_value.ActionValue`. By wrapping the outputs of Q-functions, ChainerRL can treat discrete-action Q-functions like this and [NAFs (Normalized Advantage Functions)](https://arxiv.org/abs/1603.00748) in the same way.",
"_____no_output_____"
]
],
[
[
"class QFunction(chainer.Chain):\n\n def __init__(self, obs_size, n_actions, n_hidden_channels=50):\n super().__init__(\n l0=L.Linear(obs_size, n_hidden_channels),\n l1=L.Linear(n_hidden_channels, n_hidden_channels),\n l2=L.Linear(n_hidden_channels, n_actions))\n\n def __call__(self, x, test=False):\n \"\"\"\n Args:\n x (ndarray or chainer.Variable): An observation\n test (bool): a flag indicating whether it is in test mode\n \"\"\"\n h = F.tanh(self.l0(x))\n h = F.tanh(self.l1(h))\n return chainerrl.action_value.DiscreteActionValue(self.l2(h))\n\nobs_size = env.observation_space.shape[0]\nn_actions = env.action_space.n\nq_func = QFunction(obs_size, n_actions)",
"_____no_output_____"
]
],
[
[
"If you want to use CUDA for computation, as usual as in Chainer, call `to_gpu`.",
"_____no_output_____"
]
],
[
[
"# Uncomment to use CUDA\n# q_func.to_gpu(0)",
"_____no_output_____"
]
],
[
[
"You can also use ChainerRL's predefined Q-functions.",
"_____no_output_____"
]
],
[
[
"_q_func = chainerrl.q_functions.FCStateQFunctionWithDiscreteAction(\n obs_size, n_actions,\n n_hidden_layers=2, n_hidden_channels=50)",
"_____no_output_____"
]
],
[
[
"As in Chainer, `chainer.Optimizer` is used to update models.",
"_____no_output_____"
]
],
[
[
"# Use Adam to optimize q_func. eps=1e-2 is for stability.\noptimizer = chainer.optimizers.Adam(eps=1e-2)\noptimizer.setup(q_func)",
"_____no_output_____"
]
],
[
[
"A Q-function and its optimizer are used by a DQN agent. To create a DQN agent, you need to specify a bit more parameters and configurations.",
"_____no_output_____"
]
],
[
[
"# Set the discount factor that discounts future rewards.\ngamma = 0.95\n\n# Use epsilon-greedy for exploration\nexplorer = chainerrl.explorers.ConstantEpsilonGreedy(\n epsilon=0.3, random_action_func=env.action_space.sample)\n\n# DQN uses Experience Replay.\n# Specify a replay buffer and its capacity.\nreplay_buffer = chainerrl.replay_buffer.ReplayBuffer(capacity=10 ** 6)\n\n# Since observations from CartPole-v0 is numpy.float64 while\n# Chainer only accepts numpy.float32 by default, specify\n# a converter as a feature extractor function phi.\nphi = lambda x: x.astype(np.float32, copy=False)\n\n# Now create an agent that will interact with the environment.\nagent = chainerrl.agents.DoubleDQN(\n q_func, optimizer, replay_buffer, gamma, explorer,\n replay_start_size=500, update_interval=1,\n target_update_interval=100, phi=phi)",
"_____no_output_____"
]
],
[
[
"Now you have an agent and an environment. It's time to start reinforcement learning!\n\nIn training, use `agent.act_and_train` to select exploratory actions. `agent.stop_episode_and_train` must be called after finishing an episode. You can get training statistics of the agent via `agent.get_statistics`.",
"_____no_output_____"
]
],
[
[
"n_episodes = 200\nmax_episode_len = 200\nfor i in range(1, n_episodes + 1):\n obs = env.reset()\n reward = 0\n done = False\n R = 0 # return (sum of rewards)\n t = 0 # time step\n while not done and t < max_episode_len:\n # Uncomment to watch the behaviour\n # env.render()\n action = agent.act_and_train(obs, reward)\n obs, reward, done, _ = env.step(action)\n R += reward\n t += 1\n if i % 10 == 0:\n print('episode:', i,\n 'R:', R,\n 'statistics:', agent.get_statistics())\n agent.stop_episode_and_train(obs, reward, done)\nprint('Finished.')",
"episode: 10 R: 12.0 statistics: [('average_q', 0.0077787917633448615), ('average_loss', 0)]\nepisode: 20 R: 43.0 statistics: [('average_q', 0.013923729594215806), ('average_loss', 0)]\nepisode: 30 R: 10.0 statistics: [('average_q', 0.04999595856865319), ('average_loss', 0.15626195506060395)]\nepisode: 40 R: 10.0 statistics: [('average_q', 0.18431173820404814), ('average_loss', 0.19973429628136666)]\nepisode: 50 R: 16.0 statistics: [('average_q', 0.4329778858284125), ('average_loss', 0.12129529302886367)]\nepisode: 60 R: 40.0 statistics: [('average_q', 1.5867962687319506), ('average_loss', 0.1231642400453139)]\nepisode: 70 R: 36.0 statistics: [('average_q', 4.5508317081422485), ('average_loss', 0.14574642336842872)]\nepisode: 80 R: 70.0 statistics: [('average_q', 7.293821113338115), ('average_loss', 0.222018443450522)]\nepisode: 90 R: 42.0 statistics: [('average_q', 9.706054559843952), ('average_loss', 0.22261116615911836)]\nepisode: 100 R: 148.0 statistics: [('average_q', 13.271654782141711), ('average_loss', 0.2537233644580171)]\nepisode: 110 R: 185.0 statistics: [('average_q', 17.379473389886567), ('average_loss', 0.23995480935576677)]\nepisode: 120 R: 179.0 statistics: [('average_q', 19.205810990096783), ('average_loss', 0.20982516267359438)]\nepisode: 130 R: 200.0 statistics: [('average_q', 19.86128616157245), ('average_loss', 0.17017104907517325)]\nepisode: 140 R: 160.0 statistics: [('average_q', 20.14523553965665), ('average_loss', 0.17918074812334736)]\nepisode: 150 R: 200.0 statistics: [('average_q', 20.386843352118866), ('average_loss', 0.1511973771788008)]\nepisode: 160 R: 200.0 statistics: [('average_q', 20.524274776492966), ('average_loss', 0.181143022239863)]\nepisode: 170 R: 200.0 statistics: [('average_q', 20.501493065164738), ('average_loss', 0.1426581032476842)]\nepisode: 180 R: 146.0 statistics: [('average_q', 20.37513869566722), ('average_loss', 0.12322326194384814)]\nepisode: 190 R: 55.0 statistics: [('average_q', 20.404746612680285), ('average_loss', 0.13629612704703933)]\nepisode: 200 R: 200.0 statistics: [('average_q', 20.572537269328773), ('average_loss', 0.1488116341248042)]\nFinished.\n"
]
],
[
[
"Now you finished training the agent. How good is the agent now? You can test it by using `agent.act` and `agent.stop_episode` instead. Exploration such as epsilon-greedy is not used anymore.",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n obs = env.reset()\n done = False\n R = 0\n t = 0\n while not done and t < 200:\n env.render(close=True)\n action = agent.act(obs)\n obs, r, done, _ = env.step(action)\n R += r\n t += 1\n print('test episode:', i, 'R:', R)\n agent.stop_episode()",
"test episode: 0 R: 200.0\ntest episode: 1 R: 200.0\ntest episode: 2 R: 200.0\ntest episode: 3 R: 200.0\ntest episode: 4 R: 200.0\ntest episode: 5 R: 200.0\ntest episode: 6 R: 200.0\ntest episode: 7 R: 200.0\ntest episode: 8 R: 200.0\ntest episode: 9 R: 200.0\n"
]
],
[
[
"If test scores are good enough, the only remaining task is to save the agent so that you can reuse it. What you need to do is to simply call `agent.save` to save the agent, then `agent.load` to load the saved agent.",
"_____no_output_____"
]
],
[
[
"# Save an agent to the 'agent' directory\nagent.save('agent')\n\n# Uncomment to load an agent from the 'agent' directory\n# agent.load('agent')",
"_____no_output_____"
]
],
[
[
"RL completed!\n\nBut writing code like this every time you use RL might be boring. So, ChainerRL has utility functions that do these things.",
"_____no_output_____"
]
],
[
[
"# Set up the logger to print info messages for understandability.\nimport logging\nimport sys\ngym.undo_logger_setup() # Turn off gym's default logger settings\nlogging.basicConfig(level=logging.INFO, stream=sys.stdout, format='')\n\nchainerrl.experiments.train_agent_with_evaluation(\n agent, env,\n steps=2000, # Train the agent for 2000 steps\n eval_n_runs=10, # 10 episodes are sampled for each evaluation\n max_episode_len=200, # Maximum length of each episodes\n eval_interval=1000, # Evaluate the agent after every 1000 steps\n outdir='result') # Save everything to 'result' directory",
"outdir:result step:86 episode:0 R:86.0\nstatistics:[('average_q', 20.728489019516747), ('average_loss', 0.13604925025581077)]\noutdir:result step:286 episode:1 R:200.0\nstatistics:[('average_q', 20.671014208079793), ('average_loss', 0.14984728771766473)]\noutdir:result step:396 episode:2 R:110.0\nstatistics:[('average_q', 20.658295082215886), ('average_loss', 0.16141102891913808)]\noutdir:result step:596 episode:3 R:200.0\nstatistics:[('average_q', 20.65092498811014), ('average_loss', 0.11670109444167831)]\noutdir:result step:796 episode:4 R:200.0\nstatistics:[('average_q', 20.624282196582172), ('average_loss', 0.15006617026267832)]\noutdir:result step:996 episode:5 R:200.0\nstatistics:[('average_q', 20.590381701508214), ('average_loss', 0.17453604165516437)]\noutdir:result step:1196 episode:6 R:200.0\nstatistics:[('average_q', 20.571275081196642), ('average_loss', 0.16252849495287455)]\ntest episode: 0 R: 200.0\ntest episode: 1 R: 200.0\ntest episode: 2 R: 200.0\ntest episode: 3 R: 200.0\ntest episode: 4 R: 200.0\ntest episode: 5 R: 200.0\ntest episode: 6 R: 200.0\ntest episode: 7 R: 200.0\ntest episode: 8 R: 200.0\ntest episode: 9 R: 200.0\nThe best score is updated -3.40282e+38 -> 200.0\nSaved the agent to result/1196\noutdir:result step:1244 episode:7 R:48.0\nstatistics:[('average_q', 20.44840300754298), ('average_loss', 0.1455696393507992)]\noutdir:result step:1444 episode:8 R:200.0\nstatistics:[('average_q', 20.443317168193577), ('average_loss', 0.1385756250812212)]\noutdir:result step:1644 episode:9 R:200.0\nstatistics:[('average_q', 20.388818403317572), ('average_loss', 0.11136568147911419)]\noutdir:result step:1844 episode:10 R:200.0\nstatistics:[('average_q', 20.393853468915438), ('average_loss', 0.1388451133452519)]\noutdir:result step:1951 episode:11 R:107.0\nstatistics:[('average_q', 20.403746200029968), ('average_loss', 0.1201870912602859)]\noutdir:result step:2000 episode:12 R:49.0\nstatistics:[('average_q', 20.413271961263554), ('average_loss', 0.13582760984249495)]\ntest episode: 0 R: 200.0\ntest episode: 1 R: 200.0\ntest episode: 2 R: 200.0\ntest episode: 3 R: 200.0\ntest episode: 4 R: 200.0\ntest episode: 5 R: 200.0\ntest episode: 6 R: 200.0\ntest episode: 7 R: 200.0\ntest episode: 8 R: 200.0\ntest episode: 9 R: 200.0\nSaved the agent to result/2000_finish\n"
]
],
[
[
"That's all of the ChainerRL quickstart guide. To know more about ChainerRL, please look into the `examples` directory and read and run the examples. Thank you!",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b4c4bcc41e128bafdd719378869a6aa5bd020d | 12,990 | ipynb | Jupyter Notebook | notebooks/docker_and_kubernetes/labs/2_intro_k8s.ipynb | ctivanovich/asl-ml-immersion | a2251b0368e5a5575ccdfbefd51ac1688bc7f110 | [
"Apache-2.0"
] | null | null | null | notebooks/docker_and_kubernetes/labs/2_intro_k8s.ipynb | ctivanovich/asl-ml-immersion | a2251b0368e5a5575ccdfbefd51ac1688bc7f110 | [
"Apache-2.0"
] | null | null | null | notebooks/docker_and_kubernetes/labs/2_intro_k8s.ipynb | ctivanovich/asl-ml-immersion | a2251b0368e5a5575ccdfbefd51ac1688bc7f110 | [
"Apache-2.0"
] | null | null | null | 35.108108 | 946 | 0.566436 | [
[
[
"# Introduction to Kubernetes",
"_____no_output_____"
],
[
"**Learning Objectives**\n * Create GKE cluster from command line\n * Deploy an application to your cluster\n * Cleanup, delete the cluster ",
"_____no_output_____"
],
[
"## Overview\nKubernetes is an open source project (available on [kubernetes.io](kubernetes.io)) which can run on many different environments, from laptops to high-availability multi-node clusters; from public clouds to on-premise deployments; from virtual machines to bare metal.\n\nThe goal of this lab is to provide a short introduction to Kubernetes (k8s) and some basic functionality.",
"_____no_output_____"
],
[
"## Create a GKE cluster\n\nA cluster consists of at least one cluster master machine and multiple worker machines called nodes. Nodes are Compute Engine virtual machine (VM) instances that run the Kubernetes processes necessary to make them part of the cluster.\n\n**Note**: Cluster names must start with a letter and end with an alphanumeric, and cannot be longer than 40 characters.\n\nWe'll call our cluster `asl-cluster`.",
"_____no_output_____"
]
],
[
[
"import os\n\nCLUSTER_NAME = \"asl-cluster\"\nZONE = \"us-central1-a\"\n\nos.environ[\"CLUSTER_NAME\"] = CLUSTER_NAME\nos.environ[\"ZONE\"] = ZONE",
"_____no_output_____"
]
],
[
[
"We'll set our default compute zone to `us-central1-a` and use `gcloud container clusters create ...` to create the GKE cluster. Let's first look at all the clusters we currently have. ",
"_____no_output_____"
]
],
[
[
"!gcloud container clusters list",
"NAME LOCATION MASTER_VERSION MASTER_IP MACHINE_TYPE NODE_VERSION NUM_NODES STATUS\ncluster-1 us-central1-a 1.19.14-gke.1900 35.193.84.252 custom-2-4352 1.19.14-gke.1900 2 RUNNING\n"
]
],
[
[
"**Exercise**\n\nUse `gcloud container clusters create` to create a new cluster using the `CLUSTER_NAME` we set above. This takes a few minutes...",
"_____no_output_____"
]
],
[
[
"%%bash\ngcloud container clusters create $CLUSTER_NAME --zone $ZONE",
"NAME LOCATION MASTER_VERSION MASTER_IP MACHINE_TYPE NODE_VERSION NUM_NODES STATUS\nasl-cluster us-central1-a 1.20.10-gke.1600 34.71.2.241 e2-medium 1.20.10-gke.1600 3 RUNNING\n"
]
],
[
[
"Now when we list our clusters again, we should see the cluster we created. ",
"_____no_output_____"
]
],
[
[
"!gcloud container clusters list",
"NAME LOCATION MASTER_VERSION MASTER_IP MACHINE_TYPE NODE_VERSION NUM_NODES STATUS\nasl-cluster us-central1-a 1.20.10-gke.1600 34.71.2.241 e2-medium 1.20.10-gke.1600 3 RUNNING\ncluster-1 us-central1-a 1.19.14-gke.1900 35.193.84.252 custom-2-4352 1.19.14-gke.1900 2 RUNNING\n"
]
],
[
[
"## Get authentication credentials and deploy and application\n\nAfter creating your cluster, you need authentication credentials to interact with it. Use `get-credentials` to authenticate the cluster.\n\n**Exercise**\n\nUse `gcloud container clusters get-credentials` to authenticate the cluster you created.",
"_____no_output_____"
]
],
[
[
"%%bash \ngcloud container clusters get-credentials asl-cluster --zone $ZONE",
"Fetching cluster endpoint and auth data.\nkubeconfig entry generated for asl-cluster.\n"
]
],
[
[
"You can now deploy a containerized application to the cluster. For this lab, you'll run `hello-app` in your cluster.\n\nGKE uses Kubernetes objects to create and manage your cluster's resources. Kubernetes provides the [Deployment](https://kubernetes.io/docs/concepts/workloads/controllers/deployment/) object for deploying stateless applications like web servers. [Service](https://kubernetes.io/docs/concepts/services-networking/service/) objects define rules and load balancing for accessing your application from the internet.",
"_____no_output_____"
],
[
"**Exercise**\n\nUse the `kubectl create` command to create a new Deployment `hello-server` from the `hello-app` container image. The `--image` flag to specify a container image to deploy. The `kubectl create` command pulls the example image from a Container Registry bucket. Here, use [gcr.io/google-samples/hello-app:1.0](gcr.io/google-samples/hello-app:1.0) to indicate the specific image version to pull. If a version is not specified, the latest version is used.",
"_____no_output_____"
]
],
[
[
"%%bash\nkubectl create deployment hello-server --image=gcr.io/google-samples/hello-app:1.0",
"deployment.apps/hello-server created\n"
]
],
[
[
"This Kubernetes command creates a Deployment object that represents `hello-server`. To create a Kubernetes Service, which is a Kubernetes resource that lets you expose your application to external traffic, run the `kubectl expose` command. \n\n**Exercise**\n\nUse the `kubectl expose` to expose the application. In this command, \n * `--port` specifies the port that the container exposes.\n * `type=\"LoadBalancer\"` creates a Compute Engine load balancer for your container.",
"_____no_output_____"
]
],
[
[
"%%bash\nkubectl expose deployment hello-server --type=LoadBalancer --port 8080 ",
"service/hello-server exposed\n"
]
],
[
[
"Use the `kubectl get service` command to inspect the `hello-server` Service.\n\n**Note**: It might take a minute for an external IP address to be generated. Run the previous command again if the `EXTERNAL-IP` column for `hello-server` status is pending.",
"_____no_output_____"
]
],
[
[
"!kubectl get service hello-server",
"NAME TYPE CLUSTER-IP EXTERNAL-IP PORT(S) AGE\nhello-server LoadBalancer 10.7.245.50 34.135.234.4 8080:30333/TCP 73s\n"
]
],
[
[
"You can now view the application from your web browser, open a new tab and enter the following address, replacing `EXTERNAL IP` with the EXTERNAL-IP for `hello-server`:\n\n```bash\nhttp://[EXTERNAL_IP]:8080\n```\n\nYou should see a simple page which displays\n\n```bash\nHello, world!\nVersion: 1.0.0\nHostname: hello-server-5bfd595c65-7jqkn\n```",
"_____no_output_____"
],
[
"## Cleanup\n\nDelete the cluster using `gcloud` to free up those resources. Use the `--quiet` flag if you are executing this in a notebook. Deleting the cluster can take a few minutes. ",
"_____no_output_____"
],
[
"**Exercise**\n\nDelete the cluster. Use the `--quiet` flag since we're executing in a notebook.",
"_____no_output_____"
]
],
[
[
"%%bash\nkubectl delete deployment hello-server",
"deployment.apps \"hello-server\" deleted\n"
]
],
[
[
"Copyright 2020 Google LLC Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at https://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b4e5aa68db4463858a4c1f7e2fd2cd4b74d66b | 7,110 | ipynb | Jupyter Notebook | notebooks/seldon_client.ipynb | OrthoDex/seldon-core | 1c2b511d1b14cbdae64ebde2e67f20aa18e6554e | [
"Apache-2.0"
] | 1 | 2020-03-29T18:56:31.000Z | 2020-03-29T18:56:31.000Z | notebooks/seldon_client.ipynb | OrthoDex/seldon-core | 1c2b511d1b14cbdae64ebde2e67f20aa18e6554e | [
"Apache-2.0"
] | 120 | 2020-04-27T09:48:02.000Z | 2021-07-26T06:26:10.000Z | notebooks/seldon_client.ipynb | OrthoDex/seldon-core | 1c2b511d1b14cbdae64ebde2e67f20aa18e6554e | [
"Apache-2.0"
] | 1 | 2020-03-29T18:56:33.000Z | 2020-03-29T18:56:33.000Z | 34.682927 | 778 | 0.644304 | [
[
[
"# Advanced Usage Exampes for Seldon Client",
"_____no_output_____"
],
[
"## Istio Gateway Request with token over HTTPS - no SSL verification\n\nTest against a current kubeflow cluster with Dex token authentication.\n\n 1. Install kubeflow with Dex authentication",
"_____no_output_____"
]
],
[
[
"INGRESS_HOST=!kubectl -n istio-system get service istio-ingressgateway -o jsonpath='{.status.loadBalancer.ingress[0].ip}'\nISTIO_GATEWAY=INGRESS_HOST[0]",
"_____no_output_____"
],
[
"ISTIO_GATEWAY",
"_____no_output_____"
]
],
[
[
"Get a token from the Dex gateway. At present as Dex does not support curl password credentials you will need to get it from your browser logged into the cluster. Open up a browser console and run `document.cookie`",
"_____no_output_____"
]
],
[
[
"TOKEN=\"eyJhbGciOiJIUzUxMiIsInR5cCI6IkpXVCJ9.eyJleHAiOjE1NjM2MjA0ODYsImlhdCI6MTU2MzUzNDA4NiwiaXNzIjoiMzQuNjUuNzMuMjU1IiwianRpIjoiYjllNDQxOGQtZjNmNC00NTIyLTg5ODEtNDcxOTY0ODNmODg3IiwidWlmIjoiZXlKcGMzTWlPaUpvZEhSd2N6b3ZMek0wTGpZMUxqY3pMakkxTlRvMU5UVTJMMlJsZUNJc0luTjFZaUk2SWtOcFVYZFBSMFUwVG1wbk1GbHBNV3RaYW1jMFRGUlNhVTU2VFhSUFZFSm9UMU13ZWxreVVYaE9hbGw0V21wVk1FNXFXVk5DVjNoMldUSkdjeUlzSW1GMVpDSTZJbXQxWW1WbWJHOTNMV0YxZEdoelpYSjJhV05sTFc5cFpHTWlMQ0psZUhBaU9qRTFOak0yTWpBME9EWXNJbWxoZENJNk1UVTJNelV6TkRBNE5pd2lZWFJmYUdGemFDSTZJbE5OWlZWRGJUQmFOVkZoUTNCdVNHTndRMWgwTVZFaUxDSmxiV0ZwYkNJNkltRmtiV2x1UUhObGJHUnZiaTVwYnlJc0ltVnRZV2xzWDNabGNtbG1hV1ZrSWpwMGNuVmxMQ0p1WVcxbElqb2lZV1J0YVc0aWZRPT0ifQ.7CQIz4A1s9m6lJeWTqpz_JKGArGX4e_zpRCOXXjVRJgguB3z48rSfei_KL7niMCWpruhU11c8UIw9E79PwHNNw\"",
"_____no_output_____"
]
],
[
[
"## Start Seldon Core\n\nUse the setup notebook to [Install Seldon Core](seldon_core_setup.ipynb#Install-Seldon-Core) with [Istio Ingress](seldon_core_setup.ipynb#Istio). Instructions [also online](./seldon_core_setup.html).\n\n**Note** When running helm install for this example you will need to set the istio.gateway flag to kubeflow-gateway (```--set istio.gateway=kubeflow-gateway```).",
"_____no_output_____"
]
],
[
[
"deployment_name=\"test1\"\nnamespace=\"default\"",
"_____no_output_____"
],
[
"from seldon_core.seldon_client import SeldonClient, SeldonChannelCredentials, SeldonCallCredentials\nsc = SeldonClient(deployment_name=deployment_name,namespace=namespace,gateway_endpoint=ISTIO_GATEWAY,debug=True,\n channel_credentials=SeldonChannelCredentials(verify=False),\n call_credentials=SeldonCallCredentials(token=TOKEN))",
"_____no_output_____"
],
[
"r = sc.predict(gateway=\"istio\",transport=\"rest\",shape=(1,4))\nprint(r)",
"_____no_output_____"
]
],
[
[
"Its not presently possible to use gRPC without getting access to the certificates. We will update this once its clear how to obtain them from a Kubeflow cluser setup.",
"_____no_output_____"
],
[
"## Istio - SSL Endpoint - Client Side Verification - No Authentication\n\n 1. First run through the [Istio Secure Gateway SDS example](https://istio.io/docs/tasks/traffic-management/ingress/secure-ingress-sds/) and make sure this works for you.\n * This will create certificates for `httpbin.example.com` and test them out.\n 1. Update your `/etc/hosts` file to include an entry for the ingress gateway for `httpbin.example.com` e.g. add a line like: `10.107.247.132 httpbin.example.com` replacing the ip address with your ingress gateway ip address.",
"_____no_output_____"
]
],
[
[
"# Set to folder where the httpbin certificates are\nISTIO_HTTPBIN_CERT_FOLDER='/home/clive/work/istio/httpbin.example.com'",
"_____no_output_____"
]
],
[
[
"## Start Seldon Core\n\nUse the setup notebook to [Install Seldon Core](seldon_core_setup.ipynb#Install-Seldon-Core) with [Istio Ingress](seldon_core_setup.ipynb#Istio). Instructions [also online](./seldon_core_setup.html).\n\n**Note** When running ```helm install``` for this example you will need to set the ```istio.gateway``` flag to ```mygateway``` (```--set istio.gateway=mygateway```) used in the example.",
"_____no_output_____"
]
],
[
[
"deployment_name=\"mymodel\"\nnamespace=\"default\"",
"_____no_output_____"
],
[
"from seldon_core.seldon_client import SeldonClient, SeldonChannelCredentials, SeldonCallCredentials\nsc = SeldonClient(deployment_name=deployment_name,namespace=namespace,gateway_endpoint=\"httpbin.example.com\",debug=True,\n channel_credentials=SeldonChannelCredentials(certificate_chain_file=ISTIO_HTTPBIN_CERT_FOLDER+'/2_intermediate/certs/ca-chain.cert.pem',\n root_certificates_file=ISTIO_HTTPBIN_CERT_FOLDER+'/4_client/certs/httpbin.example.com.cert.pem',\n private_key_file=ISTIO_HTTPBIN_CERT_FOLDER+'/4_client/private/httpbin.example.com.key.pem'\n ))",
"_____no_output_____"
],
[
"r = sc.predict(gateway=\"istio\",transport=\"rest\",shape=(1,4))\nprint(r)",
"_____no_output_____"
],
[
"r = sc.predict(gateway=\"istio\",transport=\"grpc\",shape=(1,4))\nprint(r)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0b4eca08bc2708e45e998732e49bdf2b529605c | 7,586 | ipynb | Jupyter Notebook | Notebooks Checkpoint/cms-queries.ipynb | aakash0017/covid-bot | 04c567e1ed5e47613e798dc2d1cf4510942839dd | [
"Apache-2.0"
] | 1 | 2021-06-07T02:54:53.000Z | 2021-06-07T02:54:53.000Z | Notebooks Checkpoint/cms-queries.ipynb | aakash0017/covid-bot | 04c567e1ed5e47613e798dc2d1cf4510942839dd | [
"Apache-2.0"
] | 2 | 2021-05-03T14:26:21.000Z | 2021-05-04T17:12:57.000Z | Notebooks Checkpoint/cms-queries.ipynb | aakash0017/covid-bot | 04c567e1ed5e47613e798dc2d1cf4510942839dd | [
"Apache-2.0"
] | 4 | 2021-04-25T08:59:37.000Z | 2021-05-11T07:50:31.000Z | 23.559006 | 194 | 0.477722 | [
[
[
"import requests\n\n# url = 'http://localhost:1337/tests'\n# myobj = {'Name': 'nidhir test1',\n# 'Email': '[email protected]',\n# 'Phoneno': 8384041898\n# }\n\n# x = requests.post(url, data = myobj, headers = {\n\n# \"Authorization\": f\"Bearer {aakash_jwt}\"}\n# )\n",
"_____no_output_____"
],
[
"aakash_jwt = \"eyJhbGciOiJIUzI1NiIsInR5cCI6IkpXVCJ9.eyJpZCI6IjYwODY1ZDUxMDZiZWQyZDhlYzZjNmJjNyIsImlhdCI6MTYxOTQxODU1OSwiZXhwIjoxNjIyMDEwNTU5fQ.Y52bTH80hazoVPomjX9jyE4LyyOKkEnZIRjAwgPeIuY\"",
"_____no_output_____"
],
[
"url = 'http://localhost:1337/tests'\nmyobj = {'Name': 'nidhir test1',\n 'Email': '[email protected]',\n 'Phoneno': 8384041898\n}\n\nx = requests.get(url, headers = {\n\n \"Authorization\": f\"Bearer {aakash_jwt}\"}\n)\n",
"_____no_output_____"
],
[
"type(x.json())",
"_____no_output_____"
],
[
"myobj = {'Name': 'nidhir test1',\n 'Email': '[email protected]',\n 'Phoneno': 8384041898\n}",
"_____no_output_____"
],
[
"res = requests.get('http://localhost:1337/data', headers = {\"Authorization\": f\"Bearer {aakash_jwt}\"})",
"_____no_output_____"
],
[
"for i in res.json():\n if i['Resources'] == 'remdisvir':\n print(i['Name'])",
"aakriti\n"
],
[
"def get_request(resource, city, endpoint, environment = 'local', url = 'http://localhost:1337/'): \n url = url + endpoint\n res = requests.get(url, headers = {\"Authorization\": f\"Bearer {aakash_jwt}\"}).json()\n resource_list = resource.split(',')\n _list = []\n for i in res:\n temp = i['Resources'].split(',')\n for j in resource_list:\n if j in temp:\n if city == i['City']:\n _list.append([i['Name'], i['City'], i['Mobile'], i['Resources']])\n\n if len(_list) == 0:\n return('Currenntly we do not have the resources please try again later')\n else: \n return _list\n\n\n",
"_____no_output_____"
],
[
"print(get_request('remdisvir,oximeter', 'delhi', 'data'))",
"[['Aakash Bhatnagar', 'delhi', '8384041898', 'oximeter, plasmaB+, oxygencylinder ']]\n"
],
[
"res.json()",
"_____no_output_____"
],
[
"dict_ = {\n 'Name': '',\n 'Email': '',\n 'City': '',\n 'State': '',\n 'Resources': '',\n 'Description': '',\n 'Mobile': '',\n}",
"_____no_output_____"
],
[
"print(dict_)",
"{'Name': '', 'Email': '', 'City': '', 'State': '', 'Resources': '', 'Description': '', 'Mobile': ''}\n"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"cd ..",
"c:\\Users\\nidbh\\work\\covid-bot\n"
],
[
"np.save('data/default_dict.npy', dict_)",
"_____no_output_____"
],
[
"np.load('data/default_dict.npy', allow_pickle=True)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b4f29894b39852f4d005979945e8e6f67b24f2 | 79,309 | ipynb | Jupyter Notebook | notebookToHtml/biosignalsnotebooks_html_publish/Categories/Install/prepare_anaconda.ipynb | biosignalsnotebooks/biosignalsnotebooks | 72b1f053320747683bb9ff123ca180cb1bd47f6a | [
"MIT"
] | 7 | 2018-11-07T14:40:13.000Z | 2019-11-03T20:38:52.000Z | notebookToHtml/biosignalsnotebooks_html_publish/Categories/Install/prepare_anaconda.ipynb | biosignalsnotebooks/biosignalsnotebooks | 72b1f053320747683bb9ff123ca180cb1bd47f6a | [
"MIT"
] | null | null | null | notebookToHtml/biosignalsnotebooks_html_publish/Categories/Install/prepare_anaconda.ipynb | biosignalsnotebooks/biosignalsnotebooks | 72b1f053320747683bb9ff123ca180cb1bd47f6a | [
"MIT"
] | 1 | 2019-06-02T07:50:41.000Z | 2019-06-02T07:50:41.000Z | 43.768764 | 5,029 | 0.522034 | [
[
[
"<table width=\"100%\">\n <tr style=\"border-bottom:solid 2pt #009EE3\">\n <td style=\"text-align:left\" width=\"10%\">\n <a href=\"prepare_anaconda.dwipynb\" download><img src=\"../../images/icons/download.png\"></a>\n </td>\n <td style=\"text-align:left\" width=\"10%\">\n <a href=\"https://mybinder.org/v2/gh/biosignalsnotebooks/biosignalsnotebooks/biosignalsnotebooks_binder?filepath=biosignalsnotebooks_environment%2Fcategories%2FInstall%2Fprepare_anaconda.dwipynb\" target=\"_blank\"><img src=\"../../images/icons/program.png\" title=\"Be creative and test your solutions !\"></a>\n </td>\n <td></td>\n <td style=\"text-align:left\" width=\"5%\">\n <a href=\"../MainFiles/biosignalsnotebooks.ipynb\"><img src=\"../../images/icons/home.png\"></a>\n </td>\n <td style=\"text-align:left\" width=\"5%\">\n <a href=\"../MainFiles/contacts.ipynb\"><img src=\"../../images/icons/contacts.png\"></a>\n </td>\n <td style=\"text-align:left\" width=\"5%\">\n <a href=\"https://github.com/biosignalsnotebooks/biosignalsnotebooks\" target=\"_blank\"><img src=\"../../images/icons/github.png\"></a>\n </td>\n <td style=\"border-left:solid 2pt #009EE3\" width=\"15%\">\n <img src=\"../../images/ost_logo.png\">\n </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"<link rel=\"stylesheet\" href=\"../../styles/theme_style.css\">\n<!--link rel=\"stylesheet\" href=\"../../styles/header_style.css\"-->\n<link rel=\"stylesheet\" href=\"https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css\">\n\n<table width=\"100%\">\n <tr>\n <td id=\"image_td\" width=\"15%\" class=\"header_image_color_13\"><div id=\"image_img\"\n class=\"header_image_13\"></div></td>\n <td class=\"header_text\"> Download, Install and Execute Anaconda </td>\n </tr>\n</table>",
"_____no_output_____"
],
[
"<div id=\"flex-container\">\n <div id=\"diff_level\" class=\"flex-item\">\n <strong>Difficulty Level:</strong> <span class=\"fa fa-star checked\"></span>\n <span class=\"fa fa-star\"></span>\n <span class=\"fa fa-star\"></span>\n <span class=\"fa fa-star\"></span>\n <span class=\"fa fa-star\"></span>\n </div>\n <div id=\"tag\" class=\"flex-item-tag\">\n <span id=\"tag_list\">\n <table id=\"tag_list_table\">\n <tr>\n <td class=\"shield_left\">Tags</td>\n <td class=\"shield_right\" id=\"tags\">install☁jupyter☁notebook☁anaconda☁download</td>\n </tr>\n </table>\n </span>\n <!-- [OR] Visit https://img.shields.io in order to create a tag badge-->\n </div>\n</div>",
"_____no_output_____"
],
[
"In every journey we always need to prepare our toolbox with the needed resources !\n\nWith <strong><span class=\"color1\">biosignalsnotebooks</span></strong> happens the same, being <strong><span class=\"color4\">Jupyter Notebook</span></strong> environment the most relevant application (that supports <strong><span class=\"color1\">biosignalsnotebooks</span></strong>) to take the maximum advantage during your learning process.\n\nIn the following sequence of instruction it will be presented the operations that should be completed in order to have <strong><span class=\"color4\">Jupyter Notebook</span></strong> ready to use and to open our <strong>ipynb</strong> files on local server.\n\n<table width=\"100%\">\n <tr>\n <td style=\"text-align:left;font-size:12pt;border-top:dotted 2px #62C3EE\">\n <span class=\"color1\">☌</span> The current <span class=\"color4\"><strong>Jupyter Notebook</strong></span> is focused on a complete Python toolbox called <a href=\"https://www.anaconda.com/distribution/\"><span class=\"color4\"><strong>Anaconda <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></span></a>.\n However, there is an alternative approach to get all things ready for starting our journey, which is described on <a href=\"../Install/prepare_jupyter.ipynb\"><span class=\"color1\"><strong>\"Download, Install and Execute Jypyter Notebook Environment\" <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></span></a>\n </td>\n </tr>\n</table>\n<hr>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<p class=\"steps\">1 - Access the <strong><span class=\"color4\">Anaconda</span></strong> official page at <a href=\"https://www.anaconda.com/distribution/\">https://www.anaconda.com/distribution/</a></p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/anaconda_page.png\">",
"_____no_output_____"
],
[
"<p class=\"steps\">2 - Click on \"Download\" button, giving a first but strong step into our final objective</p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/anaconda_download.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">3 - Specify the operating system of your local machine</p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/anaconda_download_os.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">4 - Select the version of <span class=\"color1\">Python</span> compiler to be included on <span class=\"color4\">Anaconda</span></p>\nIt is strongly advisable that you chose version <strong>3.-</strong> to ensure that all functionalities of packages like <strong><span class=\"color1\">biosignalsnotebooks</span></strong> are fully operational.",
"_____no_output_____"
],
[
"<img src=\"../../images/other/anaconda_download_version.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">5 - After defining the directory where the downloaded file will be stored, please, wait a few minutes for the end of transfer</p>\n<span class=\"color13\" style=\"font-size:30px\">⚠</span>\nThe waiting time will depend on the quality of the Internet connection !",
"_____no_output_____"
],
[
"<p class=\"steps\">6 - When download is finished navigate through your directory tree until reaching the folder where the downloaded file is located</p>\nIn our case the destination folder was <img src=\"../../images/other/anaconda_download_location.png\" style=\"display:inline;margin-top:0px\">",
"_____no_output_____"
],
[
"<p class=\"steps\">7 - Execute <span class=\"color4\">Anaconda</span> installer file with a double-click</p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/anaconda_download_installer.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">8 - Follow the sequential instructions presented on the <span class=\"color4\">Anaconda</span> installer</p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/anaconda_download_install_steps.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">9 - <span class=\"color4\">Jupyter Notebook</span> environment is included on the previous installation. For starting your first Notebook execute <span class=\"color4\">Jupyter Notebook</span></p>\nLaunch from \"Anaconda Navigator\" or through a command window, like described on the following steps.\n<p class=\"steps\">9.1 - For executing <span class=\"color4\">Jupyter Notebook</span> environment you should open a <strong>console</strong> (in your operating system).</p>\n<i>If you are a Microsoft Windows native, just type click on Windows logo (bottom-left corner of the screen) and type \"cmd\". Then press \"Enter\".</i>",
"_____no_output_____"
],
[
"<p class=\"steps\">9.2 - Type <strong>\"jupyter notebook\"</strong> inside the opened console. A local <span class=\"color4\"><strong>Jupyter Notebook</strong></span> server will be launched.</p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/open_jupyter.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">10 - Create a blank Notebook</p>\n<p class=\"steps\">10.1 - Now, you should navigate through your directories until reaching the folder where you want to create or open a Notebook (as demonstrated in the following video)</p>\n\n<span class=\"color13\" style=\"font-size:30px\">⚠</span>\n<p style=\"margin-top:0px\">You should note that your folder hierarchy is unique, so, the steps followed in the next image, will depend on your folder organisation, being merely illustrative </p>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/create_notebook_part1.gif\">",
"_____no_output_____"
],
[
"<p class=\"steps\">10.2 - For creating a new Notebook, \"New\" button (top-right zone of Jupyter Notebook interface) should be pressed and <span class=\"color1\"><strong>Python 3</strong></span> option selected.</p>\n<i>A blank Notebook will arise and now you just need to be creative and expand your thoughts to others persons!!!</i>",
"_____no_output_____"
],
[
"<img src=\"../../images/other/create_notebook_part2.gif\">",
"_____no_output_____"
],
[
"This can be the start of something great. Now you have all the software conditions to create and develop interactive tutorials, combining Python with HTML !\n\n<span class=\"color4\"><strong>Anaconda</strong></span> contains lots of additional functionalities, namely <a href=\"https://anaconda.org/anaconda/spyder\"><span class=\"color7\"><strong>Spyder <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></strong></span></a>, which is an intuitive Python editor for creating and testing your own scripts.\n\n<strong><span class=\"color7\">We hope that you have enjoyed this guide. </span><span class=\"color2\">biosignalsnotebooks</span><span class=\"color4\"> is an environment in continuous expansion, so don't stop your journey and learn more with the remaining <a href=\"../MainFiles/biosignalsnotebooks.ipynb\">Notebooks <img src=\"../../images/icons/link.png\" width=\"10px\" height=\"10px\" style=\"display:inline\"></a></span></strong> ! ",
"_____no_output_____"
],
[
"<hr>\n<table width=\"100%\">\n <tr>\n <td style=\"border-right:solid 3px #009EE3\" width=\"20%\">\n <img src=\"../../images/ost_logo.png\">\n </td>\n <td width=\"40%\" style=\"text-align:left\">\n <a href=\"../MainFiles/aux_files/biosignalsnotebooks_presentation.pdf\" target=\"_blank\">☌ Project Presentation</a>\n <br>\n <a href=\"https://github.com/biosignalsnotebooks/biosignalsnotebooks\" target=\"_blank\">☌ GitHub Repository</a>\n <br>\n <a href=\"https://pypi.org/project/biosignalsnotebooks/\" target=\"_blank\">☌ How to install biosignalsnotebooks Python package ?</a>\n <br>\n <a href=\"../MainFiles/signal_samples.ipynb\">☌ Signal Library</a>\n </td>\n <td width=\"40%\" style=\"text-align:left\">\n <a href=\"../MainFiles/biosignalsnotebooks.ipynb\">☌ Notebook Categories</a>\n <br>\n <a href=\"../MainFiles/by_diff.ipynb\">☌ Notebooks by Difficulty</a>\n <br>\n <a href=\"../MainFiles/by_signal_type.ipynb\">☌ Notebooks by Signal Type</a>\n <br>\n <a href=\"../MainFiles/by_tag.ipynb\">☌ Notebooks by Tag</a>\n </td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"from biosignalsnotebooks.__notebook_support__ import css_style_apply\ncss_style_apply()",
"_____no_output_____"
],
[
"%%html\n<script>\n // AUTORUN ALL CELLS ON NOTEBOOK-LOAD!\n require(\n ['base/js/namespace', 'jquery'],\n function(jupyter, $) {\n $(jupyter.events).on(\"kernel_ready.Kernel\", function () {\n console.log(\"Auto-running all cells-below...\");\n jupyter.actions.call('jupyter-notebook:run-all-cells-below');\n jupyter.actions.call('jupyter-notebook:save-notebook');\n });\n }\n );\n</script>",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
d0b511dd631a106423823ed350d10578a71462b2 | 81,782 | ipynb | Jupyter Notebook | code/4_basic_3d_GAN/1_main_code/train_3dgan.ipynb | vmos1/cosmogan_pytorch | 75d3d4f652a92d45d823a051b750b35d802e2317 | [
"BSD-3-Clause-LBNL"
] | 1 | 2020-10-19T18:52:50.000Z | 2020-10-19T18:52:50.000Z | code/4_basic_3d_GAN/1_main_code/train_3dgan.ipynb | vmos1/cosmogan_pytorch | 75d3d4f652a92d45d823a051b750b35d802e2317 | [
"BSD-3-Clause-LBNL"
] | 1 | 2020-11-13T22:35:02.000Z | 2020-11-14T02:00:44.000Z | code/4_basic_3d_GAN/1_main_code/train_3dgan.ipynb | vmos1/cosmogan_pytorch | 75d3d4f652a92d45d823a051b750b35d802e2317 | [
"BSD-3-Clause-LBNL"
] | null | null | null | 52.933333 | 1,765 | 0.534592 | [
[
[
"# Testing cosmogan\nApril 19, 2021\n\nBorrowing pieces of code from : \n\n- https://github.com/pytorch/tutorials/blob/11569e0db3599ac214b03e01956c2971b02c64ce/beginner_source/dcgan_faces_tutorial.py\n- https://github.com/exalearn/epiCorvid/tree/master/cGAN",
"_____no_output_____"
]
],
[
[
"import os\nimport random\nimport logging\nimport sys\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.parallel\nimport torch.backends.cudnn as cudnn\nimport torch.optim as optim\nimport torch.utils.data\nfrom torchsummary import summary\nfrom torch.utils.data import DataLoader, TensorDataset\nimport torch.distributed as dist\nfrom torch.nn.parallel import DistributedDataParallel\n# import torch.fft\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib.animation as animation\n# from IPython.display import HTML\n\nimport argparse\nimport time\nfrom datetime import datetime\nimport glob\nimport pickle\nimport yaml\nimport collections\nimport socket\nimport shutil\n\n# # Import modules from other files\n# from utils import *\n# from spec_loss import *",
"_____no_output_____"
],
[
"%matplotlib widget",
"_____no_output_____"
]
],
[
[
"## Modules",
"_____no_output_____"
]
],
[
[
"### Transformation functions for image pixel values\ndef f_transform(x):\n return 2.*x/(x + 4.) - 1.\n\ndef f_invtransform(s):\n return 4.*(1. + s)/(1. - s)\n\n \n# Generator Code\nclass View(nn.Module):\n def __init__(self, shape):\n super(View, self).__init__()\n self.shape = shape\n\n def forward(self, x):\n return x.view(*self.shape)\n\ndef f_get_model(gdict):\n ''' Module to define Generator and Discriminator'''\n\n if gdict['image_size']==64:\n\n class Generator(nn.Module):\n def __init__(self, gdict):\n super(Generator, self).__init__()\n\n ## Define new variables from dict\n keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']\n ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())\n\n self.main = nn.Sequential(\n # nn.ConvTranspose3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)\n nn.Linear(nz,nc*ngf*8**3),# 262144\n nn.BatchNorm3d(nc,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n View(shape=[-1,ngf*8,4,4,4]),\n nn.ConvTranspose3d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),\n nn.BatchNorm3d(ngf*4,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n # state size. (ngf*4) x 8 x 8\n nn.ConvTranspose3d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),\n nn.BatchNorm3d(ngf*2,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n # state size. (ngf*2) x 16 x 16\n nn.ConvTranspose3d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),\n nn.BatchNorm3d(ngf,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n # state size. (ngf) x 32 x 32\n nn.ConvTranspose3d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),\n nn.Tanh()\n )\n\n def forward(self, ip):\n return self.main(ip)\n\n class Discriminator(nn.Module):\n def __init__(self, gdict):\n super(Discriminator, self).__init__()\n\n ## Define new variables from dict\n keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']\n ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values()) \n\n self.main = nn.Sequential(\n # input is (nc) x 64 x 64 x 64\n # nn.Conv3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)\n nn.Conv3d(nc, ndf,kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf) x 32 x 32\n nn.Conv3d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*2) x 16 x 16\n nn.Conv3d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*4) x 8 x 8\n nn.Conv3d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*8) x 4 x 4\n nn.Flatten(),\n nn.Linear(nc*ndf*8*8*8, 1)\n # nn.Sigmoid()\n )\n\n def forward(self, ip):\n # print(ip.shape)\n results=[ip]\n lst_idx=[]\n for i,submodel in enumerate(self.main.children()):\n mid_output=submodel(results[-1])\n results.append(mid_output)\n ## Select indices in list corresponding to output of Conv layers\n if submodel.__class__.__name__.startswith('Conv'):\n # print(submodel.__class__.__name__)\n # print(mid_output.shape)\n lst_idx.append(i)\n\n FMloss=True\n if FMloss:\n ans=[results[1:][i] for i in lst_idx + [-1]]\n else :\n ans=results[-1]\n return ans\n\n elif gdict['image_size']==128:\n\n class Generator(nn.Module):\n def __init__(self, gdict):\n super(Generator, self).__init__()\n\n ## Define new variables from dict\n keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']\n ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())\n\n self.main = nn.Sequential(\n # nn.ConvTranspose3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)\n nn.Linear(nz,nc*ngf*8**3*8),# 262144\n nn.BatchNorm3d(nc,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n View(shape=[-1,ngf*8,8,8,8]),\n nn.ConvTranspose3d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),\n nn.BatchNorm3d(ngf*4,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n # state size. (ngf*4) x 8 x 8\n nn.ConvTranspose3d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),\n nn.BatchNorm3d(ngf*2,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n # state size. (ngf*2) x 16 x 16\n nn.ConvTranspose3d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),\n nn.BatchNorm3d(ngf,eps=1e-05, momentum=0.9, affine=True),\n nn.ReLU(inplace=True),\n # state size. (ngf) x 32 x 32\n nn.ConvTranspose3d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),\n nn.Tanh()\n )\n\n def forward(self, ip):\n return self.main(ip)\n\n class Discriminator(nn.Module):\n def __init__(self, gdict):\n super(Discriminator, self).__init__()\n\n ## Define new variables from dict\n keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']\n ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values()) \n\n self.main = nn.Sequential(\n # input is (nc) x 64 x 64 x 64\n # nn.Conv3d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)\n nn.Conv3d(nc, ndf,kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf) x 32 x 32\n nn.Conv3d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*2) x 16 x 16\n nn.Conv3d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*4) x 8 x 8\n nn.Conv3d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),\n nn.BatchNorm3d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),\n nn.LeakyReLU(0.2, inplace=True),\n # state size. (ndf*8) x 4 x 4\n nn.Flatten(),\n nn.Linear(nc*ndf*8*8*8*8, 1)\n # nn.Sigmoid()\n )\n\n def forward(self, ip):\n results=[ip]\n lst_idx=[]\n for i,submodel in enumerate(self.main.children()):\n mid_output=submodel(results[-1])\n results.append(mid_output)\n ## Select indices in list corresponding to output of Conv layers\n if submodel.__class__.__name__.startswith('Conv'):\n # print(submodel.__class__.__name__)\n # print(mid_output.shape)\n lst_idx.append(i)\n\n FMloss=True\n if FMloss:\n ans=[results[1:][i] for i in lst_idx + [-1]]\n else :\n ans=results[-1]\n return ans\n \n return Generator, Discriminator\n\n\ndef f_gen_images(gdict,netG,optimizerG,ip_fname,op_loc,op_strg='inf_img_',op_size=500):\n '''Generate images for best saved models\n Arguments: gdict, netG, optimizerG, \n ip_fname: name of input file\n op_strg: [string name for output file]\n op_size: Number of images to generate\n '''\n\n nz,device=gdict['nz'],gdict['device']\n\n try:# handling cpu vs gpu\n if torch.cuda.is_available(): checkpoint=torch.load(ip_fname)\n else: checkpoint=torch.load(ip_fname,map_location=torch.device('cpu'))\n except Exception as e:\n print(e)\n print(\"skipping generation of images for \",ip_fname)\n return\n \n ## Load checkpoint\n if gdict['multi-gpu']:\n netG.module.load_state_dict(checkpoint['G_state'])\n else:\n netG.load_state_dict(checkpoint['G_state'])\n \n ## Load other stuff\n iters=checkpoint['iters']\n epoch=checkpoint['epoch']\n optimizerG.load_state_dict(checkpoint['optimizerG_state_dict'])\n \n # Generate batch of latent vectors\n noise = torch.randn(op_size, 1, 1, 1, nz, device=device)\n # Generate fake image batch with G\n netG.eval() ## This is required before running inference\n with torch.no_grad(): ## This is important. fails without it for multi-gpu\n gen = netG(noise)\n gen_images=gen.detach().cpu().numpy()\n print(gen_images.shape)\n \n op_fname='%s_epoch-%s_step-%s.npy'%(op_strg,epoch,iters)\n np.save(op_loc+op_fname,gen_images)\n\n print(\"Image saved in \",op_fname)\n \ndef f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,netG,netD,optimizerG,optimizerD,save_loc):\n ''' Checkpoint model '''\n \n if gdict['multi-gpu']: ## Dataparallel\n torch.save({'epoch':epoch,'iters':iters,'best_chi1':best_chi1,'best_chi2':best_chi2,\n 'G_state':netG.module.state_dict(),'D_state':netD.module.state_dict(),'optimizerG_state_dict':optimizerG.state_dict(),\n 'optimizerD_state_dict':optimizerD.state_dict()}, save_loc) \n else :\n torch.save({'epoch':epoch,'iters':iters,'best_chi1':best_chi1,'best_chi2':best_chi2,\n 'G_state':netG.state_dict(),'D_state':netD.state_dict(),'optimizerG_state_dict':optimizerG.state_dict(),\n 'optimizerD_state_dict':optimizerD.state_dict()}, save_loc)\n \ndef f_load_checkpoint(ip_fname,netG,netD,optimizerG,optimizerD,gdict):\n ''' Load saved checkpoint\n Also loads step, epoch, best_chi1, best_chi2'''\n \n print(\"torch device\",torch.device('cuda',torch.cuda.current_device()))\n \n try:\n checkpoint=torch.load(ip_fname,map_location=torch.device('cuda',torch.cuda.current_device()))\n except Exception as e:\n print(\"Error loading saved checkpoint\",ip_fname)\n print(e)\n raise SystemError\n \n ## Load checkpoint\n if gdict['multi-gpu']:\n netG.module.load_state_dict(checkpoint['G_state'])\n netD.module.load_state_dict(checkpoint['D_state'])\n else:\n netG.load_state_dict(checkpoint['G_state'])\n netD.load_state_dict(checkpoint['D_state'])\n \n optimizerD.load_state_dict(checkpoint['optimizerD_state_dict'])\n optimizerG.load_state_dict(checkpoint['optimizerG_state_dict'])\n \n iters=checkpoint['iters']\n epoch=checkpoint['epoch']\n best_chi1=checkpoint['best_chi1']\n best_chi2=checkpoint['best_chi2']\n\n netG.train()\n netD.train()\n \n return iters,epoch,best_chi1,best_chi2,netD,optimizerD,netG,optimizerG\n",
"_____no_output_____"
],
[
"####################\n### Pytorch code ###\n####################\n\ndef f_get_rad(img):\n ''' Get the radial tensor for use in f_torch_get_azimuthalAverage '''\n \n height,width,depth=img.shape[-3:]\n # Create a grid of points with x and y and z coordinates\n z,y,x = np.indices([height,width,depth])\n \n center=[]\n if not center:\n center = np.array([(x.max()-x.min())/2.0, (y.max()-y.min())/2.0, (z.max()-z.min())/2.0])\n\n # Get the radial coordinate for every grid point. Array has the shape of image\n r= torch.tensor(np.sqrt((x-center[0])**2 + (y-center[1])**2 + (z-center[2])**2))\n \n # Get sorted radii\n ind = torch.argsort(torch.reshape(r, (-1,)))\n\n return r.detach(),ind.detach()\n\n\ndef f_torch_get_azimuthalAverage(image,r,ind):\n \"\"\"\n Calculate the azimuthally averaged radial profile.\n\n image - The 2D image\n center - The [x,y] pixel coordinates used as the center. The default is \n None, which then uses the center of the image (including \n fracitonal pixels).\n source: https://www.astrobetter.com/blog/2010/03/03/fourier-transforms-of-images-in-python/\n \"\"\"\n \n# height, width = image.shape\n# # Create a grid of points with x and y coordinates\n# y, x = np.indices([height,width])\n\n# if not center:\n# center = np.array([(x.max()-x.min())/2.0, (y.max()-y.min())/2.0])\n\n# # Get the radial coordinate for every grid point. Array has the shape of image\n# r = torch.tensor(np.hypot(x - center[0], y - center[1]))\n\n# # Get sorted radii\n# ind = torch.argsort(torch.reshape(r, (-1,)))\n\n r_sorted = torch.gather(torch.reshape(r, ( -1,)),0, ind)\n i_sorted = torch.gather(torch.reshape(image, ( -1,)),0, ind)\n \n # Get the integer part of the radii (bin size = 1)\n r_int=r_sorted.to(torch.int32)\n\n # Find all pixels that fall within each radial bin.\n deltar = r_int[1:] - r_int[:-1] # Assumes all radii represented\n rind = torch.reshape(torch.where(deltar)[0], (-1,)) # location of changes in radius\n nr = (rind[1:] - rind[:-1]).type(torch.float) # number of radius bin\n\n # Cumulative sum to figure out sums for each radius bin\n \n csum = torch.cumsum(i_sorted, axis=-1)\n tbin = torch.gather(csum, 0, rind[1:]) - torch.gather(csum, 0, rind[:-1])\n radial_prof = tbin / nr\n\n return radial_prof\n\ndef f_torch_fftshift(real, imag):\n for dim in range(0, len(real.size())):\n real = torch.roll(real, dims=dim, shifts=real.size(dim)//2)\n imag = torch.roll(imag, dims=dim, shifts=imag.size(dim)//2)\n return real, imag\n\ndef f_torch_compute_spectrum(arr,r,ind):\n \n GLOBAL_MEAN=1.0\n arr=(arr-GLOBAL_MEAN)/(GLOBAL_MEAN)\n \n y1=torch.rfft(arr,signal_ndim=3,onesided=False)\n real,imag=f_torch_fftshift(y1[:,:,:,0],y1[:,:,:,1]) ## last index is real/imag part ## Mod for 3D\n \n# # For pytorch 1.8\n# y1=torch.fft.fftn(arr,dim=(-3,-2,-1))\n# real,imag=f_torch_fftshift(y1.real,y1.imag) \n \n y2=real**2+imag**2 ## Absolute value of each complex number\n z1=f_torch_get_azimuthalAverage(y2,r,ind) ## Compute radial profile\n return z1\n\ndef f_torch_compute_batch_spectrum(arr,r,ind):\n \n batch_pk=torch.stack([f_torch_compute_spectrum(i,r,ind) for i in arr])\n \n return batch_pk\n\ndef f_torch_image_spectrum(x,num_channels,r,ind):\n '''\n Data has to be in the form (batch,channel,x,y)\n '''\n mean=[[] for i in range(num_channels)] \n var=[[] for i in range(num_channels)] \n\n for i in range(num_channels):\n arr=x[:,i,:,:,:] # Mod for 3D\n batch_pk=f_torch_compute_batch_spectrum(arr,r,ind)\n mean[i]=torch.mean(batch_pk,axis=0)\n# var[i]=torch.std(batch_pk,axis=0)/np.sqrt(batch_pk.shape[0])\n# var[i]=torch.std(batch_pk,axis=0)\n var[i]=torch.var(batch_pk,axis=0)\n \n mean=torch.stack(mean)\n var=torch.stack(var)\n \n if (torch.isnan(mean).any() or torch.isnan(var).any()):\n print(\"Nans in spectrum\",mean,var)\n if torch.isnan(x).any():\n print(\"Nans in Input image\")\n\n return mean,var\n\ndef f_compute_hist(data,bins):\n \n try: \n hist_data=torch.histc(data,bins=bins)\n ## A kind of normalization of histograms: divide by total sum\n hist_data=(hist_data*bins)/torch.sum(hist_data)\n except Exception as e:\n print(e)\n hist_data=torch.zeros(bins)\n\n return hist_data\n\n### Losses \ndef loss_spectrum(spec_mean,spec_mean_ref,spec_var,spec_var_ref,image_size,lambda_spec_mean,lambda_spec_var):\n ''' Loss function for the spectrum : mean + variance \n Log(sum( batch value - expect value) ^ 2 )) '''\n \n if (torch.isnan(spec_mean).any() or torch.isnan(spec_var).any()):\n ans=torch.tensor(float(\"inf\"))\n return ans\n \n idx=int(image_size/2) ### For the spectrum, use only N/2 indices for loss calc.\n ### Warning: the first index is the channel number.For multiple channels, you are averaging over them, which is fine.\n \n loss_mean=torch.log(torch.mean(torch.pow(spec_mean[:,:idx]-spec_mean_ref[:,:idx],2)))\n loss_var=torch.log(torch.mean(torch.pow(spec_var[:,:idx]-spec_var_ref[:,:idx],2)))\n \n ans=lambda_spec_mean*loss_mean+lambda_spec_var*loss_var\n \n if (torch.isnan(ans).any()) : \n print(\"loss spec mean %s, loss spec var %s\"%(loss_mean,loss_var))\n print(\"spec mean %s, ref %s\"%(spec_mean, spec_mean_ref))\n print(\"spec var %s, ref %s\"%(spec_var, spec_var_ref))\n# raise SystemExit\n \n return ans\n \ndef loss_hist(hist_sample,hist_ref):\n \n lambda1=1.0\n return lambda1*torch.log(torch.mean(torch.pow(hist_sample-hist_ref,2)))\n\ndef f_FM_loss(real_output,fake_output,lambda_fm,gdict):\n '''\n Module to implement Feature-Matching loss. Reads all but last elements of Discriminator ouput\n '''\n FM=torch.Tensor([0.0]).to(gdict['device'])\n for i,j in zip(real_output[:-1],fake_output[:-1]):\n# print(i.shape,j.shape)\n real_mean=torch.mean(i)\n fake_mean=torch.mean(j)\n# print(real_mean,fake_mean)\n FM=FM.clone()+torch.sum(torch.square(real_mean-fake_mean))\n return lambda_fm*FM\n\ndef f_gp_loss(grads,l=1.0):\n '''\n Module to implement gradient penalty loss.\n '''\n loss=torch.mean(torch.sum(torch.square(grads),dim=[1,2,3]))\n return l*loss",
"_____no_output_____"
]
],
[
[
"## Train loop",
"_____no_output_____"
]
],
[
[
"### Train code ###\ndef f_train_loop(gan_model,Dset,metrics_df,gdict,fixed_noise):\n ''' Train epochs '''\n ## Define new variables from dict\n keys=['image_size','start_epoch','epochs','iters','best_chi1','best_chi2','save_dir','device','flip_prob','nz','batch_size','bns']\n image_size,start_epoch,epochs,iters,best_chi1,best_chi2,save_dir,device,flip_prob,nz,batchsize,bns=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())\n \n for epoch in range(start_epoch,epochs):\n t_epoch_start=time.time()\n for count, data in enumerate(Dset.train_dataloader):\n\n ####### Train GAN ########\n gan_model.netG.train(); gan_model.netD.train(); ### Need to add these after inference and before training\n\n tme1=time.time()\n ### Update D network: maximize log(D(x)) + log(1 - D(G(z)))\n gan_model.netD.zero_grad()\n\n real_cpu = data[0].to(device)\n real_cpu.requires_grad=True\n b_size = real_cpu.size(0)\n real_label = torch.full((b_size,), 1, device=device,dtype=float)\n fake_label = torch.full((b_size,), 0, device=device,dtype=float)\n g_label = torch.full((b_size,), 1, device=device,dtype=float) ## No flipping for Generator labels\n # Flip labels with probability flip_prob\n for idx in np.random.choice(np.arange(b_size),size=int(np.ceil(b_size*flip_prob))):\n real_label[idx]=0; fake_label[idx]=1\n\n # Generate fake image batch with G\n noise = torch.randn(b_size, 1, 1, 1, nz, device=device) ### Mod for 3D\n fake = gan_model.netG(noise) \n\n # Forward pass real batch through D\n real_output = gan_model.netD(real_cpu)\n errD_real = gan_model.criterion(real_output[-1].view(-1), real_label.float())\n errD_real.backward(retain_graph=True)\n D_x = real_output[-1].mean().item()\n\n # Forward pass fake batch through D\n fake_output = gan_model.netD(fake.detach()) # The detach is important\n errD_fake = gan_model.criterion(fake_output[-1].view(-1), fake_label.float())\n errD_fake.backward(retain_graph=True)\n D_G_z1 = fake_output[-1].mean().item()\n \n errD = errD_real + errD_fake \n\n if gdict['lambda_gp']: ## Add gradient - penalty loss\n grads=torch.autograd.grad(outputs=real_output[-1],inputs=real_cpu,grad_outputs=torch.ones_like(real_output[-1]),allow_unused=False,create_graph=True)[0]\n gp_loss=f_gp_loss(grads,gdict['lambda_gp'])\n gp_loss.backward(retain_graph=True)\n errD = errD + gp_loss\n else:\n gp_loss=torch.Tensor([np.nan])\n \n if gdict['grad_clip']:\n nn.utils.clip_grad_norm_(gan_model.netD.parameters(),gdict['grad_clip'])\n\n gan_model.optimizerD.step()\n lr_d=gan_model.optimizerD.param_groups[0]['lr']\n gan_model.schedulerD.step()\n \n# dict_keys(['train_data_loader', 'r', 'ind', 'train_spec_mean', 'train_spec_var', 'train_hist', 'val_spec_mean', 'val_spec_var', 'val_hist'])\n\n ###Update G network: maximize log(D(G(z)))\n gan_model.netG.zero_grad()\n output = gan_model.netD(fake)\n errG_adv = gan_model.criterion(output[-1].view(-1), g_label.float())\n# errG_adv.backward(retain_graph=True)\n # Histogram pixel intensity loss\n hist_gen=f_compute_hist(fake,bins=bns)\n hist_loss=loss_hist(hist_gen,Dset.train_hist.to(device))\n\n # Add spectral loss\n mean,var=f_torch_image_spectrum(f_invtransform(fake),1,Dset.r.to(device),Dset.ind.to(device))\n spec_loss=loss_spectrum(mean,Dset.train_spec_mean.to(device),var,Dset.train_spec_var.to(device),image_size,gdict['lambda_spec_mean'],gdict['lambda_spec_var'])\n\n errG=errG_adv\n if gdict['lambda_spec_mean']: \n# spec_loss.backward(retain_graph=True)\n errG = errG+ spec_loss \n if gdict['lambda_fm']:## Add feature matching loss\n fm_loss=f_FM_loss([i.detach() for i in real_output],output,gdict['lambda_fm'],gdict)\n# fm_loss.backward(retain_graph=True)\n errG= errG+ fm_loss\n else: \n fm_loss=torch.Tensor([np.nan])\n\n if torch.isnan(errG).any():\n logging.info(errG)\n raise SystemError\n \n # Calculate gradients for G\n errG.backward()\n D_G_z2 = output[-1].mean().item()\n \n ### Implement Gradient clipping\n if gdict['grad_clip']:\n nn.utils.clip_grad_norm_(gan_model.netG.parameters(),gdict['grad_clip'])\n \n gan_model.optimizerG.step()\n lr_g=gan_model.optimizerG.param_groups[0]['lr']\n gan_model.schedulerG.step()\n \n tme2=time.time()\n ####### Store metrics ########\n # Output training stats\n if gdict['world_rank']==0:\n if ((count % gdict['checkpoint_size'] == 0)):\n logging.info('[%d/%d][%d/%d]\\tLoss_D: %.4f\\tLoss_adv: %.4f\\tLoss_G: %.4f\\tD(x): %.4f\\tD(G(z)): %.4f / %.4f'\n % (epoch, epochs, count, len(Dset.train_dataloader), errD.item(), errG_adv.item(),errG.item(), D_x, D_G_z1, D_G_z2)),\n logging.info(\"Spec loss: %s,\\t hist loss: %s\"%(spec_loss.item(),hist_loss.item())),\n logging.info(\"Training time for step %s : %s\"%(iters, tme2-tme1))\n\n # Save metrics\n cols=['step','epoch','Dreal','Dfake','Dfull','G_adv','G_full','spec_loss','hist_loss','fm_loss','gp_loss','D(x)','D_G_z1','D_G_z2','lr_d','lr_g','time']\n vals=[iters,epoch,errD_real.item(),errD_fake.item(),errD.item(),errG_adv.item(),errG.item(),spec_loss.item(),hist_loss.item(),fm_loss.item(),gp_loss.item(),D_x,D_G_z1,D_G_z2,lr_d,lr_g,tme2-tme1]\n for col,val in zip(cols,vals): metrics_df.loc[iters,col]=val\n\n ### Checkpoint the best model\n checkpoint=True\n iters += 1 ### Model has been updated, so update iters before saving metrics and model.\n\n ### Compute validation metrics for updated model\n gan_model.netG.eval()\n with torch.no_grad():\n fake = gan_model.netG(fixed_noise)\n hist_gen=f_compute_hist(fake,bins=bns)\n hist_chi=loss_hist(hist_gen,Dset.val_hist.to(device))\n mean,var=f_torch_image_spectrum(f_invtransform(fake),1,Dset.r.to(device),Dset.ind.to(device))\n spec_chi=loss_spectrum(mean,Dset.val_spec_mean.to(device),var,Dset.val_spec_var.to(device),image_size,gdict['lambda_spec_mean'],gdict['lambda_spec_var'])\n\n # Storing chi for next step\n for col,val in zip(['spec_chi','hist_chi'],[spec_chi.item(),hist_chi.item()]): metrics_df.loc[iters,col]=val \n\n # Checkpoint model for continuing run\n if count == len(Dset.train_dataloader)-1: ## Check point at last step of epoch\n f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_last.tar') \n\n if (checkpoint and (epoch > 1)): # Choose best models by metric\n if hist_chi< best_chi1:\n f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_best_hist.tar')\n best_chi1=hist_chi.item()\n logging.info(\"Saving best hist model at epoch %s, step %s.\"%(epoch,iters))\n\n if spec_chi< best_chi2:\n f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_best_spec.tar')\n best_chi2=spec_chi.item()\n logging.info(\"Saving best spec model at epoch %s, step %s\"%(epoch,iters))\n\n# if (iters in gdict['save_steps_list']) :\n if ((gdict['save_steps_list']=='all') and (iters % gdict['checkpoint_size'] == 0)):\n f_save_checkpoint(gdict,epoch,iters,best_chi1,best_chi2,gan_model.netG,gan_model.netD,gan_model.optimizerG,gan_model.optimizerD,save_loc=save_dir+'/models/checkpoint_{0}.tar'.format(iters))\n logging.info(\"Saving given-step at epoch %s, step %s.\"%(epoch,iters))\n\n # Save G's output on fixed_noise\n if ((iters % gdict['checkpoint_size'] == 0) or ((epoch == epochs-1) and (count == len(Dset.train_dataloader)-1))):\n gan_model.netG.eval()\n with torch.no_grad():\n fake = gan_model.netG(fixed_noise).detach().cpu()\n img_arr=np.array(fake)\n fname='gen_img_epoch-%s_step-%s'%(epoch,iters)\n np.save(save_dir+'/images/'+fname,img_arr)\n \n t_epoch_end=time.time()\n if gdict['world_rank']==0:\n logging.info(\"Time taken for epoch %s, count %s: %s for rank %s\"%(epoch,count,t_epoch_end-t_epoch_start,gdict['world_rank']))\n # Save Metrics to file after each epoch\n metrics_df.to_pickle(save_dir+'/df_metrics.pkle')\n logging.info(\"best chis: {0}, {1}\".format(best_chi1,best_chi2))\n\n",
"_____no_output_____"
]
],
[
[
"## Start",
"_____no_output_____"
]
],
[
[
"### Setup modules ###\ndef f_manual_add_argparse():\n ''' use only in jpt notebook'''\n args=argparse.Namespace()\n args.config='config_3dgan_128_cori.yaml'\n args.mode='fresh'\n args.local_rank=0\n args.facility='cori'\n args.distributed=False\n\n# args.mode='continue'\n \n return args\n\ndef f_parse_args():\n \"\"\"Parse command line arguments.Only for .py file\"\"\"\n parser = argparse.ArgumentParser(description=\"Run script to train GAN using pytorch\", formatter_class=argparse.ArgumentDefaultsHelpFormatter)\n add_arg = parser.add_argument\n \n add_arg('--config','-cfile', type=str, default='config_3d_Cgan.yaml', help='Name of config file')\n add_arg('--mode','-m', type=str, choices=['fresh','continue','fresh_load'],default='fresh', help='Whether to start fresh run or continue previous run or fresh run loading a config file.')\n add_arg(\"--local_rank\", default=0, type=int,help='Local rank of GPU on node. Using for pytorch DDP. ')\n add_arg(\"--facility\", default='cori', choices=['cori','summit'],type=str,help='Facility: cori or summit ')\n add_arg(\"--ddp\", dest='distributed' ,default=False,action='store_true',help='use Distributed DataParallel for Pytorch or DataParallel')\n \n return parser.parse_args()\n\ndef try_barrier(rank):\n \"\"\"\n Used in Distributed data parallel\n Attempt a barrier but ignore any exceptions\n \"\"\"\n print('BAR %d'%rank)\n try:\n dist.barrier()\n except:\n pass\n\ndef f_init_gdict(args,gdict):\n ''' Create global dictionary gdict from args and config file'''\n \n ## read config file\n config_file=args.config\n with open(config_file) as f:\n config_dict= yaml.load(f, Loader=yaml.SafeLoader)\n \n gdict=config_dict['parameters']\n\n args_dict=vars(args)\n ## Add args variables to gdict\n for key in args_dict.keys():\n gdict[key]=args_dict[key]\n\n if gdict['distributed']: \n assert not gdict['lambda_gp'],\"GP couplings is %s. Cannot use Gradient penalty loss in pytorch DDP\"%(gdict['lambda_gp'])\n else : print(\"Not using DDP\")\n return gdict\n\n\ndef f_get_img_samples(ip_arr,rank=0,num_ranks=1):\n '''\n Module to get part of the numpy image file\n '''\n \n data_size=ip_arr.shape[0]\n size=data_size//num_ranks\n \n if gdict['batch_size']>size:\n print(\"Caution: batchsize %s is greater than samples per GPU %s\"%(gdict['batch_size'],size))\n raise SystemExit\n \n ### Get a set of random indices from numpy array\n random=False\n if random:\n idxs=np.arange(ip_arr.shape[0])\n np.random.shuffle(idxs)\n rnd_idxs=idxs[rank*(size):(rank+1)*size]\n arr=ip_arr[rnd_idxs].copy()\n \n else: arr=ip_arr[rank*(size):(rank+1)*size].copy()\n \n return arr\n\ndef f_setup(gdict,metrics_df,log):\n ''' \n Set up directories, Initialize random seeds, add GPU info, add logging info.\n '''\n \n torch.backends.cudnn.benchmark=True\n# torch.autograd.set_detect_anomaly(True)\n\n ## New additions. Code taken from Jan B.\n os.environ['MASTER_PORT'] = \"8885\"\n\n if gdict['facility']=='summit':\n get_master = \"echo $(cat {} | sort | uniq | grep -v batch | grep -v login | head -1)\".format(os.environ['LSB_DJOB_HOSTFILE'])\n os.environ['MASTER_ADDR'] = str(subprocess.check_output(get_master, shell=True))[2:-3]\n os.environ['WORLD_SIZE'] = os.environ['OMPI_COMM_WORLD_SIZE']\n os.environ['RANK'] = os.environ['OMPI_COMM_WORLD_RANK']\n gdict['local_rank'] = int(os.environ['OMPI_COMM_WORLD_LOCAL_RANK'])\n else:\n if gdict['distributed']:\n os.environ['WORLD_SIZE'] = os.environ['SLURM_NTASKS']\n os.environ['RANK'] = os.environ['SLURM_PROCID']\n gdict['local_rank'] = int(os.environ['SLURM_LOCALID'])\n\n ## Special declarations\n gdict['ngpu']=torch.cuda.device_count()\n gdict['device']=torch.device(\"cuda\" if (torch.cuda.is_available()) else \"cpu\")\n gdict['multi-gpu']=True if (gdict['device'].type == 'cuda') and (gdict['ngpu'] > 1) else False \n \n ########################\n ###### Set up Distributed Data parallel ######\n if gdict['distributed']:\n# gdict['local_rank']=args.local_rank ## This is needed when using pytorch -m torch.distributed.launch\n gdict['world_size']=int(os.environ['WORLD_SIZE'])\n torch.cuda.set_device(gdict['local_rank']) ## Very important\n dist.init_process_group(backend='nccl', init_method=\"env://\") \n gdict['world_rank']= dist.get_rank()\n \n device = torch.cuda.current_device()\n logging.info(\"World size %s, world rank %s, local rank %s device %s, hostname %s, GPUs on node %s\\n\"%(gdict['world_size'],gdict['world_rank'],gdict['local_rank'],device,socket.gethostname(),gdict['ngpu']))\n \n # Divide batch size by number of GPUs\n# gdict['batch_size']=gdict['batch_size']//gdict['world_size']\n else:\n gdict['world_size'],gdict['world_rank'],gdict['local_rank']=1,0,0\n \n ########################\n ###### Set up directories #######\n ### sync up so that time is the same for each GPU for DDP\n if gdict['mode'] in ['fresh','fresh_load']:\n ### Create prefix for foldername \n if gdict['world_rank']==0: ### For rank=0, create directory name string and make directories\n dt_strg=datetime.now().strftime('%Y%m%d_%H%M%S') ## time format\n dt_lst=[int(i) for i in dt_strg.split('_')] # List storing day and time \n dt_tnsr=torch.LongTensor(dt_lst).to(gdict['device']) ## Create list to pass to other GPUs \n\n else: dt_tnsr=torch.Tensor([0,0]).long().to(gdict['device'])\n ### Pass directory name to other ranks\n if gdict['distributed']: dist.broadcast(dt_tnsr, src=0)\n\n gdict['save_dir']=gdict['op_loc']+str(int(dt_tnsr[0]))+'_'+str(int(dt_tnsr[1]))+'_'+gdict['run_suffix']\n \n if gdict['world_rank']==0: # Create directories for rank 0\n ### Create directories\n if not os.path.exists(gdict['save_dir']):\n os.makedirs(gdict['save_dir']+'/models')\n os.makedirs(gdict['save_dir']+'/images')\n shutil.copy(gdict['config'],gdict['save_dir']) \n \n elif gdict['mode']=='continue': ## For checkpointed runs\n gdict['save_dir']=gdict['ip_fldr']\n ### Read loss data\n metrics_df=pd.read_pickle(gdict['save_dir']+'/df_metrics.pkle').astype(np.float64)\n \n ########################\n ### Initialize random seed\n \n manualSeed = np.random.randint(1, 10000) if gdict['seed']=='random' else int(gdict['seed'])\n# print(\"Seed\",manualSeed,gdict['world_rank'])\n random.seed(manualSeed)\n np.random.seed(manualSeed)\n torch.manual_seed(manualSeed)\n torch.cuda.manual_seed_all(manualSeed)\n \n if gdict['deterministic']:\n logging.info(\"Running with deterministic sequence. Performance will be slower\")\n torch.backends.cudnn.deterministic=True\n# torch.backends.cudnn.enabled = False\n torch.backends.cudnn.benchmark = False \n \n ########################\n if log:\n ### Write all logging.info statements to stdout and log file\n logfile=gdict['save_dir']+'/log.log'\n if gdict['world_rank']==0:\n logging.basicConfig(level=logging.DEBUG, filename=logfile, filemode=\"a+\", format=\"%(asctime)-15s %(levelname)-8s %(message)s\")\n\n Lg = logging.getLogger()\n Lg.setLevel(logging.DEBUG)\n lg_handler_file = logging.FileHandler(logfile)\n lg_handler_stdout = logging.StreamHandler(sys.stdout)\n Lg.addHandler(lg_handler_file)\n Lg.addHandler(lg_handler_stdout)\n\n logging.info('Args: {0}'.format(args))\n logging.info('Start: %s'%(datetime.now().strftime('%Y-%m-%d %H:%M:%S')))\n \n if gdict['distributed']: try_barrier(gdict['world_rank'])\n\n if gdict['world_rank']!=0:\n logging.basicConfig(level=logging.DEBUG, filename=logfile, filemode=\"a+\", format=\"%(asctime)-15s %(levelname)-8s %(message)s\")\n\n return metrics_df\n\nclass Dataset:\n def __init__(self,gdict):\n '''\n Load training dataset and compute spectrum and histogram for a small batch of training and validation dataset.\n '''\n ## Load training dataset\n t0a=time.time()\n img=np.load(gdict['ip_fname'],mmap_mode='r')[:gdict['num_imgs']]\n # print(\"Shape of input file\",img.shape)\n img=f_get_img_samples(img,gdict['world_rank'],gdict['world_size']) \n\n t_img=torch.from_numpy(img)\n dataset=TensorDataset(t_img)\n self.train_dataloader=DataLoader(dataset,batch_size=gdict['batch_size'],shuffle=True,num_workers=0,drop_last=True)\n logging.info(\"Size of dataset for GPU %s : %s\"%(gdict['world_rank'],len(self.train_dataloader.dataset)))\n\n t0b=time.time()\n logging.info(\"Time for creating dataloader\",t0b-t0a,gdict['world_rank'])\n \n # Precompute spectrum and histogram for small training and validation data for computing losses\n with torch.no_grad():\n val_img=np.load(gdict['ip_fname'],mmap_mode='r')[-100:].copy()\n t_val_img=torch.from_numpy(val_img).to(gdict['device'])\n # Precompute radial coordinates\n r,ind=f_get_rad(val_img)\n self.r,self.ind=r.to(gdict['device']),ind.to(gdict['device'])\n\n # Compute\n self.train_spec_mean,self.train_spec_var=f_torch_image_spectrum(f_invtransform(t_val_img),1,self.r,self.ind)\n self.train_hist=f_compute_hist(t_val_img,bins=gdict['bns'])\n \n # Repeat for validation dataset\n val_img=np.load(gdict['ip_fname'],mmap_mode='r')[-200:-100].copy()\n t_val_img=torch.from_numpy(val_img).to(gdict['device'])\n \n # Compute\n self.val_spec_mean,self.val_spec_var=f_torch_image_spectrum(f_invtransform(t_val_img),1,self.r,self.ind)\n self.val_hist=f_compute_hist(t_val_img,bins=gdict['bns'])\n del val_img; del t_val_img; del img; del t_img;\n\nclass GAN_model():\n def __init__(self,gdict,print_model=False):\n \n def weights_init(m):\n '''custom weights initialization called on netG and netD '''\n classname = m.__class__.__name__\n if classname.find('Conv') != -1:\n nn.init.normal_(m.weight.data, 0.0, 0.02)\n elif classname.find('BatchNorm') != -1:\n nn.init.normal_(m.weight.data, 1.0, 0.02)\n nn.init.constant_(m.bias.data, 0)\n \n ## Choose model\n Generator, Discriminator=f_get_model(gdict) ## Mod for cGAN\n \n # Create Generator\n self.netG = Generator(gdict).to(gdict['device'])\n self.netG.apply(weights_init)\n # Create Discriminator\n self.netD = Discriminator(gdict).to(gdict['device'])\n self.netD.apply(weights_init)\n\n if print_model:\n if gdict['world_rank']==0:\n print(self.netG)\n # summary(netG,(1,1,64))\n print(self.netD)\n # summary(netD,(1,128,128))\n print(\"Number of GPUs used %s\"%(gdict['ngpu']))\n\n if (gdict['multi-gpu']):\n if not gdict['distributed']:\n self.netG = nn.DataParallel(self.netG, list(range(gdict['ngpu'])))\n self.netD = nn.DataParallel(self.netD, list(range(gdict['ngpu'])))\n else:\n self.netG=DistributedDataParallel(self.netG,device_ids=[gdict['local_rank']],output_device=[gdict['local_rank']])\n self.netD=DistributedDataParallel(self.netD,device_ids=[gdict['local_rank']],output_device=[gdict['local_rank']])\n\n #### Initialize networks ####\n # self.criterion = nn.BCELoss()\n self.criterion = nn.BCEWithLogitsLoss()\n\n self.optimizerD = optim.Adam(self.netD.parameters(), lr=gdict['learn_rate_d'], betas=(gdict['beta1'], 0.999),eps=1e-7)\n self.optimizerG = optim.Adam(self.netG.parameters(), lr=gdict['learn_rate_g'], betas=(gdict['beta1'], 0.999),eps=1e-7)\n \n if gdict['distributed']: try_barrier(gdict['world_rank'])\n\n if gdict['mode']=='fresh':\n iters,start_epoch,best_chi1,best_chi2=0,0,1e10,1e10 \n \n elif gdict['mode']=='continue':\n iters,start_epoch,best_chi1,best_chi2,self.netD,self.optimizerD,self.netG,self.optimizerG=f_load_checkpoint(gdict['save_dir']+'/models/checkpoint_last.tar',\\\n self.netG,self.netD,self.optimizerG,self.optimizerD,gdict) \n if gdict['world_rank']==0: logging.info(\"\\nContinuing existing run. Loading checkpoint with epoch {0} and step {1}\\n\".format(start_epoch,iters))\n if gdict['distributed']: try_barrier(gdict['world_rank'])\n start_epoch+=1 ## Start with the next epoch \n \n elif gdict['mode']=='fresh_load':\n iters,start_epoch,best_chi1,best_chi2,self.netD,self.optimizerD,self.netG,self.optimizerG=f_load_checkpoint(gdict['chkpt_file'],\\\n self.netG,self.netD,self.optimizerG,self.optimizerD,gdict) \n if gdict['world_rank']==0: logging.info(\"Fresh run loading checkpoint file {0}\".format(gdict['chkpt_file']))\n# if gdict['distributed']: try_barrier(gdict['world_rank'])\n iters,start_epoch,best_chi1,best_chi2=0,0,1e10,1e10 \n \n ## Add to gdict\n for key,val in zip(['best_chi1','best_chi2','iters','start_epoch'],[best_chi1,best_chi2,iters,start_epoch]): gdict[key]=val\n \n ## Set up learn rate scheduler\n lr_stepsize=int((gdict['num_imgs'])/(gdict['batch_size']*gdict['world_size'])) # convert epoch number to step \n lr_d_epochs=[i*lr_stepsize for i in gdict['lr_d_epochs']] \n lr_g_epochs=[i*lr_stepsize for i in gdict['lr_g_epochs']]\n self.schedulerD = optim.lr_scheduler.MultiStepLR(self.optimizerD, milestones=lr_d_epochs,gamma=gdict['lr_d_gamma'])\n self.schedulerG = optim.lr_scheduler.MultiStepLR(self.optimizerG, milestones=lr_g_epochs,gamma=gdict['lr_g_gamma'])\n",
"_____no_output_____"
]
],
[
[
"## Main",
"_____no_output_____"
]
],
[
[
"#########################\n### Main code #######\n#########################\n\nif __name__==\"__main__\":\n jpt=False\n jpt=True ##(different for jupyter notebook)\n t0=time.time()\n t0=time.time()\n args=f_parse_args() if not jpt else f_manual_add_argparse()\n\n #################################\n ### Set up global dictionary###\n gdict={}\n gdict=f_init_gdict(args,gdict)\n# gdict['num_imgs']=200\n\n if jpt: ## override for jpt nbks\n gdict['num_imgs']=400\n gdict['run_suffix']='nb_test'\n \n ### Set up metrics dataframe\n cols=['step','epoch','Dreal','Dfake','Dfull','G_adv','G_full','spec_loss','hist_loss','spec_chi','hist_chi','gp_loss','fm_loss','D(x)','D_G_z1','D_G_z2','time']\n metrics_df=pd.DataFrame(columns=cols)\n \n # Setup\n metrics_df=f_setup(gdict,metrics_df,log=(not jpt))\n \n ## Build GAN\n gan_model=GAN_model(gdict,False)\n fixed_noise = torch.randn(gdict['op_size'], 1, 1, 1, gdict['nz'], device=gdict['device']) #Latent vectors to view G progress # Mod for 3D\n\n if gdict['distributed']: try_barrier(gdict['world_rank'])\n\n ## Load data and precompute\n Dset=Dataset(gdict)\n \n #################################\n ########## Train loop and save metrics and images ######\n if gdict['distributed']: try_barrier(gdict['world_rank'])\n\n if gdict['world_rank']==0: \n logging.info(gdict)\n logging.info(\"Starting Training Loop...\")\n \n f_train_loop(gan_model,Dset,metrics_df,gdict,fixed_noise)\n \n if gdict['world_rank']==0: ## Generate images for best saved models ######\n op_loc=gdict['save_dir']+'/images/'\n ip_fname=gdict['save_dir']+'/models/checkpoint_best_spec.tar'\n f_gen_images(gdict,gan_model.netG,gan_model.optimizerG,ip_fname,op_loc,op_strg='best_spec',op_size=32)\n ip_fname=gdict['save_dir']+'/models/checkpoint_best_hist.tar'\n f_gen_images(gdict,gan_model.netG,gan_model.optimizerG,ip_fname,op_loc,op_strg='best_hist',op_size=32)\n \n tf=time.time()\n logging.info(\"Total time %s\"%(tf-t0))\n logging.info('End: %s'%(datetime.now().strftime('%Y-%m-%d %H:%M:%S')))\n",
"Not using DDP\n"
],
[
"# metrics_df.plot('step','time')\nmetrics_df",
"_____no_output_____"
],
[
"gan_model.optimizerG.param_groups[0]['lr']\n# metrics_df['lr_d']\n",
"_____no_output_____"
],
[
"# summary(gan_model.netG,(1,1,64))\nsummary(gan_model.netD,(1,128,128,128))",
"----------------------------------------------------------------\n Layer (type) Output Shape Param #\n================================================================\n Conv3d-1 [-1, 64, 64, 64, 64] 8,064\n BatchNorm3d-2 [-1, 64, 64, 64, 64] 128\n LeakyReLU-3 [-1, 64, 64, 64, 64] 0\n Conv3d-4 [-1, 128, 32, 32, 32] 1,024,128\n BatchNorm3d-5 [-1, 128, 32, 32, 32] 256\n LeakyReLU-6 [-1, 128, 32, 32, 32] 0\n Conv3d-7 [-1, 256, 16, 16, 16] 4,096,256\n BatchNorm3d-8 [-1, 256, 16, 16, 16] 512\n LeakyReLU-9 [-1, 256, 16, 16, 16] 0\n Conv3d-10 [-1, 512, 8, 8, 8] 16,384,512\n BatchNorm3d-11 [-1, 512, 8, 8, 8] 1,024\n LeakyReLU-12 [-1, 512, 8, 8, 8] 0\n Flatten-13 [-1, 262144] 0\n Linear-14 [-1, 1] 262,145\n================================================================\nTotal params: 21,777,025\nTrainable params: 21,777,025\nNon-trainable params: 0\n----------------------------------------------------------------\nInput size (MB): 8.00\nForward/backward pass size (MB): 512.00\nParams size (MB): 83.07\nEstimated Total Size (MB): 603.07\n----------------------------------------------------------------\n"
],
[
"# gdict",
"_____no_output_____"
]
],
[
[
"### Debug",
"_____no_output_____"
]
],
[
[
"# class Generator(nn.Module):\n# def __init__(self, gdict):\n# super(Generator, self).__init__()\n\n# ## Define new variables from dict\n# keys=['ngpu','nz','nc','ngf','kernel_size','stride','g_padding']\n# ngpu, nz,nc,ngf,kernel_size,stride,g_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values())\n\n# self.main = nn.Sequential(\n# # nn.ConvTranspose2d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)\n# nn.Linear(nz,nc*ngf*8*8*8),# 32768\n# nn.BatchNorm2d(nc,eps=1e-05, momentum=0.9, affine=True),\n# nn.ReLU(inplace=True),\n# View(shape=[-1,ngf*8,8,8]),\n# nn.ConvTranspose2d(ngf * 8, ngf * 4, kernel_size, stride, g_padding, output_padding=1, bias=False),\n# nn.BatchNorm2d(ngf*4,eps=1e-05, momentum=0.9, affine=True),\n# nn.ReLU(inplace=True),\n# # state size. (ngf*4) x 8 x 8\n# nn.ConvTranspose2d( ngf * 4, ngf * 2, kernel_size, stride, g_padding, 1, bias=False),\n# nn.BatchNorm2d(ngf*2,eps=1e-05, momentum=0.9, affine=True),\n# nn.ReLU(inplace=True),\n# # state size. (ngf*2) x 16 x 16\n# nn.ConvTranspose2d( ngf * 2, ngf, kernel_size, stride, g_padding, 1, bias=False),\n# nn.BatchNorm2d(ngf,eps=1e-05, momentum=0.9, affine=True),\n# nn.ReLU(inplace=True),\n# # state size. (ngf) x 32 x 32\n# nn.ConvTranspose2d( ngf, nc, kernel_size, stride,g_padding, 1, bias=False),\n# nn.Tanh()\n# )\n \n# def forward(self, ip):\n# return self.main(ip)\n\n# class Discriminator(nn.Module):\n# def __init__(self, gdict):\n# super(Discriminator, self).__init__()\n \n# ## Define new variables from dict\n# keys=['ngpu','nz','nc','ndf','kernel_size','stride','d_padding']\n# ngpu, nz,nc,ndf,kernel_size,stride,d_padding=list(collections.OrderedDict({key:gdict[key] for key in keys}).values()) \n\n# self.main = nn.Sequential(\n# # input is (nc) x 64 x 64\n# # nn.Conv2d(in_channels, out_channels, kernel_size,stride,padding,output_padding,groups,bias, Dilation,padding_mode)\n# nn.Conv2d(nc, ndf,kernel_size, stride, d_padding, bias=True),\n# nn.BatchNorm2d(ndf,eps=1e-05, momentum=0.9, affine=True),\n# nn.LeakyReLU(0.2, inplace=True),\n# # state size. (ndf) x 32 x 32\n# nn.Conv2d(ndf, ndf * 2, kernel_size, stride, d_padding, bias=True),\n# nn.BatchNorm2d(ndf * 2,eps=1e-05, momentum=0.9, affine=True),\n# nn.LeakyReLU(0.2, inplace=True),\n# # state size. (ndf*2) x 16 x 16\n# nn.Conv2d(ndf * 2, ndf * 4, kernel_size, stride, d_padding, bias=True),\n# nn.BatchNorm2d(ndf * 4,eps=1e-05, momentum=0.9, affine=True),\n# nn.LeakyReLU(0.2, inplace=True),\n# # state size. (ndf*4) x 8 x 8\n# nn.Conv2d(ndf * 4, ndf * 8, kernel_size, stride, d_padding, bias=True),\n# nn.BatchNorm2d(ndf * 8,eps=1e-05, momentum=0.9, affine=True),\n# nn.LeakyReLU(0.2, inplace=True),\n# # state size. (ndf*8) x 4 x 4\n# nn.Flatten(),\n# nn.Linear(nc*ndf*8*8*8, 1)\n# # nn.Sigmoid()\n# )\n\n# def forward(self, ip):\n# # print(ip.shape)\n# results=[ip]\n# lst_idx=[]\n# for i,submodel in enumerate(self.main.children()):\n# mid_output=submodel(results[-1])\n# results.append(mid_output)\n# ## Select indices in list corresponding to output of Conv layers\n# if submodel.__class__.__name__.startswith('Conv'):\n# # print(submodel.__class__.__name__)\n# # print(mid_output.shape)\n# lst_idx.append(i)\n\n# FMloss=True\n# if FMloss:\n# ans=[results[1:][i] for i in lst_idx + [-1]]\n# else :\n# ans=results[-1]\n# return ans\n\n",
"_____no_output_____"
],
[
"# netG = Generator(gdict).to(gdict['device'])\n# netG.apply(weights_init)\n# # # # print(netG)\n# # summary(netG,(1,1,64))\n# # Create Discriminator\n# netD = Discriminator(gdict).to(gdict['device'])\n# netD.apply(weights_init)\n# # print(netD)\n# summary(netD,(1,128,128))",
"_____no_output_____"
],
[
"# noise = torch.randn(gdict['batchsize'], 1, 1, gdict['nz'], device=gdict['device'])\n# fake = netG(noise) \n# # Forward pass real batch through D\n# output = netD(fake)\n# print([i.shape for i in output])",
"_____no_output_____"
],
[
"0.5**10",
"_____no_output_____"
],
[
"70000/(8*6*8)",
"_____no_output_____"
],
[
"gdict.keys()",
"_____no_output_____"
],
[
"for key in ['batch_size','num_imgs','ngpu']:\n print(key,gdict[key])",
"batch_size 4\nnum_imgs 40\nngpu 1\n"
],
[
"gdict['world_size']",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b5166f52e4c35665ed51ca09c08535b30c8d6f | 910,872 | ipynb | Jupyter Notebook | notebooks/p2c4/0.4-imad-cluster-analysis.ipynb | imad24/pc_clustering | b76cb5a0e6429610802594cc1ca5218dfcdb3605 | [
"MIT"
] | null | null | null | notebooks/p2c4/0.4-imad-cluster-analysis.ipynb | imad24/pc_clustering | b76cb5a0e6429610802594cc1ca5218dfcdb3605 | [
"MIT"
] | null | null | null | notebooks/p2c4/0.4-imad-cluster-analysis.ipynb | imad24/pc_clustering | b76cb5a0e6429610802594cc1ca5218dfcdb3605 | [
"MIT"
] | 1 | 2019-05-27T21:34:35.000Z | 2019-05-27T21:34:35.000Z | 187.460794 | 83,170 | 0.799765 | [
[
[
"# Exploring Clustering Results\nThe file containing the clustering results is stored in the processed data folder with the suffix clean. The index is set to the first __Product group key__.\n\nAs a reminder the file is organized in three columns: _Product Group Key_, _Cluster Number_ and the corresponding _Centroid_ of the cluster.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\n# add the 'src' directory as one where we can import modules\nroot_dir = os.path.join(os.getcwd(),os.pardir,os.pardir)\n\nimport pandas as pd\nimport math\nimport numpy as np\n\nimport pandas as pd\nimport numpy as np\n\nimport matplotlib.pyplot as plt\nimport copy as cp\n\nimport seaborn as sns\n\nimport statsmodels.api as sm\n\n\nfrom IPython.display import display\n\nraw_path = os.path.join(root_dir,\"data\\\\raw\\\\\")\ninterim_path = os.path.join(root_dir,\"data\\\\interim\\\\\") \nprocessed_path = os.path.join(root_dir,\"data\\\\processed\\\\\")\n\n\nreports_path = os.path.join(root_dir,\"reports\\\\\")\nmodels_path = os.path.join(root_dir,\"models\\\\\")\n\n\nfile_name = \"euc_p2_clustering_clean_mois_v2.csv\"\n\ndf_prd_cluster = pd.read_csv(models_path+file_name, sep=';', encoding='utf-8').drop('Unnamed: 0',axis=1).set_index('Product')\n\nprint(df_prd_cluster.shape)\ndf_prd_cluster.head()\n",
"(382, 2)\n"
]
],
[
[
"## Get clients description",
"_____no_output_____"
]
],
[
[
"file_name1 = \"data_client_bnd_ita.csv\"\nfile_name2 = \"data_client_bnd_ita2.csv\"\n\nnon_unique_features = [\"Key_lvl1\",\"Key_lvl2\",\"Key_lvl3\",\"CONCESS PDL\",\"CLIENTE FATT\",\"CONCESS FATT\",\"PTF SPEDIZIONE\",\"TYPE_DISTRIB\"]\n\nunique_features = [\"Key_lvl4\",\"Key_lvl5\"]\n\n\nclient_df1 = pd.read_csv(raw_path+file_name1, sep=';', encoding='iso8859_2').fillna(\"NA\")[unique_features]\\\n .drop_duplicates()\n \nclient_df2 = pd.read_csv(raw_path+file_name2, sep=';', encoding='iso8859_2').fillna(\"NA\")[unique_features]\\\n .drop_duplicates()\n \n \nclient_df = pd.concat([client_df1,client_df2], axis=0, ignore_index=True, copy=True).drop_duplicates()\nprint(client_df1.shape)\nprint(client_df2.shape)\nprint(client_df.shape)\n\nm = pd.merge(client_df1.iloc[:,:1],client_df2.iloc[:,:1],how='outer',on=['Key_lvl4'],indicator='both').drop_duplicates() \ndif = m[m['both']!='both'].reset_index(drop=True)\ndisplay(dif.head())",
"_____no_output_____"
]
],
[
[
"## Get Products description\nIn order to get the product features description, an inner join on the product group key is operated on the cluster result with the products description file.\n\nSince the clustering was calculated on the second level group, some columns of the description file must be dropped in order to avoid duplicates of the first level products (mainly Promo and Standard version of the products)",
"_____no_output_____"
]
],
[
[
"file_name1 = \"bnd_products_desc.csv\"\nfile_name2 = \"bnd_products_desc2.csv\"\n\nnon_unique_features=[\"Key\",\"Description\",\"CONFEZIONE\",\\\n \"CONFEZIONE (Description)\",\"IMBALLO\",\"STD/PROMO\",\"IMBALLO (Description)\",\"STD/PROMO (Description)\",\\\n \"TIPO ARTICOLO\",\"TIPO ARTICOLO (Description)\"]\n\ncode_features = [\"FAM DETTAGLIATA\",\"FAM AGGREGATA\",\"MARCHIO\",\"GRUPPO MARCHIO\",\"PACKAGING\",\"SOTTO-TECNO\",\\\n \"PRODOTTO\",\"CANALE DISTRIB\",\"CLASSE COGE\",\"FAM MARKETING\",\"BIOLOGICO\",\"GRUPPO MARCA COGE\",\"Product Group key6\"]\n\nunbalanced = [\"CANALE DISTRIB (Description)\",\"CLASSE COGE (Description)\",\"BIOLOGICO (Description)\"]\n\ndf_produit1 = pd.read_csv(raw_path+file_name1, sep=';', encoding='iso8859_2')\\\n .drop(non_unique_features,axis=1)\\\n .drop(code_features,axis=1)\\\n .drop_duplicates()\\\n .dropna().reset_index(drop=True).apply(lambda x:x.astype(str).str.upper())\n \n \ndf_produit2 = pd.read_csv(raw_path+file_name2, sep=';', encoding='iso8859_2')\\\n .drop(non_unique_features,axis=1)\\\n .drop(code_features,axis=1)\\\n .drop_duplicates()\\\n .dropna().reset_index(drop=True).apply(lambda x:x.astype(str).str.upper())\n \n\nm = pd.merge(df_produit1.iloc[:,:1],df_produit2.iloc[:,:1],how='outer',on=['Product Group key'],indicator='both').drop_duplicates() \ndif = m[m['both']!='both'].reset_index(drop=True)\n\n# df_produit = pd.concat([df_produit1,df_produit2], axis=0, ignore_index=True, copy=True)\\\n# .drop_duplicates([\"Product Group key\",\"Product Group key2\"])\n\n\n#df_produit1.to_csv(interim_path+\"\\\\unique\\\\bnd_products_desc1.csv\",sep=';',encoding='iso8859_2',index=False)\n#df_produit2.to_csv(interim_path+\"\\\\unique\\\\bnd_products_desc2.csv\",sep=';',encoding='iso8859_2',index=False)\n#df_produit.to_csv(interim_path+\"\\\\unique\\\\bnd_products_desc.csv\",sep=';',encoding='iso8859_2',index=False)\n\n\ndf_produit = df_produit2.drop_duplicates([\"Product Group key\"])\n#Remove XX products weird\nmask_XX = df_produit[\"Product Group key3\"].str.endswith(\"XXX\")\ndf_produit = df_produit[~mask_XX]\n\n\n\n#Join with clusters\nproduct_cluster = df_produit.join(df_prd_cluster,on='Product Group key',how='inner').reset_index(drop = True)\nprint(product_cluster.shape)\nproduct_cluster.head()\n#product_cluster.to_csv(interim_path+\"\\\\unique\\\\bnd_product_cluster.csv\",sep=';',encoding='iso8859_2',index=False)",
"(382, 19)\n"
],
[
"display(df[[\"Product Group key2\"]].drop_duplicates())\ndisplay(df[[\"Product Group key3\"]].drop_duplicates())\ndisplay(df[[\"Product Group key4\"]].drop_duplicates())\n\n",
"_____no_output_____"
]
],
[
[
"## Merge Products and Clients tables",
"_____no_output_____"
]
],
[
[
"# produit_client_cluster = pd.merge(product_cluster,client_df,how='inner', left_on=[\"Client\"],right_on =[\"Key_lvl4\"] )#.drop([\"Key_lvl4\"],axis=1)\n\n\n\n# clusters = produit_client_cluster[\"Cluster\"]\n# centroids = produit_client_cluster[\"Centroid\"]\n# produit_client_cluster = produit_client_cluster.drop([\"Cluster\",\"Centroid\"],axis=1)\n\n# pos = len(produit_client_cluster.columns)\n# produit_client_cluster.insert(pos,\"Cluster\",clusters)\n# produit_client_cluster.insert(pos+1,\"Centroid\",centroids)\n\n\n# all_features = produit_client_cluster.columns[:-2].drop(unbalanced)\n\n\n\n# print(produit_client_cluster.shape)\n# produit_client_cluster.tail()\n\n\n# absent = produit_client_cluster[produit_client_cluster[\"Key_lvl5\"].isnull()][\"Client\"].drop_duplicates()\n# absent.tail()\n\n\n\n\nall_features = product_cluster.columns[:-2]\n",
"_____no_output_____"
]
],
[
[
"Save the final result into a csv file for further exploration",
"_____no_output_____"
]
],
[
[
"filename = 'bnd_product_cluster_clean.csv'\nfile_name = \"p2_clustering_clean_mois.csv\"\nproduct_cluster.to_csv(processed_path+filename,sep=';',encoding='iso8859_2')\n",
"_____no_output_____"
]
],
[
[
"# Homogeneity Test\nIn order to detect specific caraterstics for each resulted cluster we perform a statistic test based on Pearsons chi-square score with the hypothesis of a uniform distribution.\n\nFeatures with the pvalues lower than 0.1 are displayed for analysis",
"_____no_output_____"
]
],
[
[
"def cramer_v(chisq,n,k,r=1):\n return math.sqrt(chisq/(n * min(k-1,r-1) ))",
"_____no_output_____"
]
],
[
[
"## Calculate modalities frequency through clusters\nAs a first step, all the distrubtions of modalities across features and clusters are calculated and stored in one array structered as follows:\n\nOne array for each cluster which contains a dictionnary of features. Each feature is again a dictionary of modalities and their occurence in that cluster",
"_____no_output_____"
]
],
[
[
"#get the features\nfeatures = all_features\n\n#get the clusters (actually its a range(1,nb_cluster))\nclusters = set(product_cluster['Cluster'].values)\n\n#array to store each cluster and freq for all the features\nclusters_feature_dist = [0] #to shift the indices to clusters\n\n#loop trhough features\n\nfor c in clusters:\n feature_dist = dict()\n for feature in features:\n freq = product_cluster[product_cluster['Cluster']==c].groupby(feature)[feature].count()\n feature_dist[feature]=freq.to_dict()\n clusters_feature_dist.append(feature_dist)\n\n",
"_____no_output_____"
]
],
[
[
"## Chi-square test over clusters",
"_____no_output_____"
]
],
[
[
"from scipy.stats import chisquare\n\npthreashold = 0.2\n\n#get the features\nfeatures = all_features\n\nclusters = [6]\n\n\nres_features_over_cluster = [0]\nfor c in clusters:\n #align each feature with its distrubtion in this cluster c\n cluster_feature_dist = clusters_feature_dist[c]\n dist = [len(x) for x in list(cluster_feature_dist.values())]\n keys = list(cluster_feature_dist.keys()) \n\n #plot the dist of number of elements by feature in this clust\n plt.title(\"Feature distribution in the cluster %d\"%c)\n plt.bar(range(len(keys)),dist)\n plt.xticks(range(len(keys)),keys,rotation=70)\n \n #for each feature display its distribution over modalities\n for feature in features:\n #get information from the previous array\n cluster_feature_dist = clusters_feature_dist[c]\n feature_distribution = list(cluster_feature_dist[feature].values())\n feature_keys = list(cluster_feature_dist[feature].keys())\n nftrs = len(feature_keys)\n chisq, p = chisquare(feature_distribution)\n if p<pthreashold:\n plt.figure()\n plt.title(\"%s modalities distribution - pvalue = %.9f\"%(feature,p))\n plt.bar(np.arange(nftrs),feature_distribution)\n plt.xticks(np.arange(nftrs)+(1.0/nftrs),feature_keys,rotation=70 if nftrs>5 else 0)\n plt.show(block = True)\n \n",
"_____no_output_____"
]
],
[
[
"## Calculate modalities frequency through features",
"_____no_output_____"
]
],
[
[
"#get the features\nfeatures = all_features\n\n#get the clusters (actually its a range(1,nb_cluster))\nclusters = set(product_cluster['Cluster'].values)\n\n#dict to store each feater and freq for all the clusters\nfeatures_clust_dist = dict()\n\n#invert the dict and get it by feature \nfor f in features:\n freq = dict()\n for c in clusters: \n freq[c] = clusters_feature_dist[c][f]\n features_clust_dist[f] = freq",
"_____no_output_____"
]
],
[
[
"## Chi-square test over features",
"_____no_output_____"
]
],
[
[
"pthreashold = 0.2\nclusters = set(product_cluster['Cluster'].values)\n\nfeatures = all_features\nfeatures = [\"FAM MARKETING (Description)\"]\n\nfor f in features:\n for c in clusters:\n #get information from the previous array\n feature_clust_dist = features_clust_dist[f]\n feature_distribution = list(feature_clust_dist[c].values())\n feature_keys = list(feature_clust_dist[c].keys())\n nftrs = len(feature_keys)\n chisq, p = chisquare(feature_distribution)\n if p<pthreashold:\n plt.figure()\n plt.title(\"%s: Cluster %d distribution - pvalue = %.9f\"%(f,c,p))\n plt.bar(np.arange(nftrs),feature_distribution)\n plt.xticks(np.arange(nftrs)+(1.0/nftrs),feature_keys,rotation=70 if nftrs>5 else 0)\n \n plt.show(block = True) \n\n",
"_____no_output_____"
]
],
[
[
"## Modalities distribution",
"_____no_output_____"
]
],
[
[
"clusters = set(product_cluster['Cluster'].values)\nnclusters = len(clusters)\n#get the features\nfeatures = product_cluster.columns[0:-2]\nfeatures = features.drop(unbalanced)\n\n\nmodalities_clust_dist = dict()\n\nfor f in features:\n feature_sum=[]\n modalities = set(product_cluster[f].values)\n modalities_distribution=dict()\n for m in modalities:\n modality_distribution = np.zeros((nclusters+1))\n for c in clusters:\n #get information from the previous array\n feature_clust_dist = features_clust_dist[f]\n modality_distribution[c] +=(feature_clust_dist[c][m] if m in feature_clust_dist[c] else 0)\n modalities_distribution[m] = modality_distribution \n modalities_clust_dist[f] = modalities_distribution ",
"_____no_output_____"
]
],
[
[
"## Chi-square test for modalities over clusters",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nclusters = set(product_cluster['Cluster'].values)\nnclusters = len(clusters)\n\n\npthreashold = 0.2\n\nn_min_dist = 1\nmin_members = 5\n\n\nmin_dust = True\nfor f in features:\n modalities = set(product_cluster[f].values)\n r = len(modalities)\n for m in modalities:\n modality_dist = modalities_clust_dist[f][m]\n md = np.count_nonzero(modality_dist)<=n_min_dist and np.max(modality_dist)>min_members\n chisq, p = chisquare(modality_dist)\n if p<pthreashold and (md and min_dust):\n plt.figure()\n plt.title(\"%s: %s Distribution - pvalue = %.9f\"%(f,m,p))\n plt.bar(np.arange(nclusters)+1,modality_dist[1:])\n plt.xticks(np.arange(nclusters)+(1.0/nclusters)+1,np.arange(nclusters)+1,rotation=90,size=8)\n if np.max(modality_dist[1:])<10: plt.ylim(0,10)\n plt.show(block = True) ",
"_____no_output_____"
]
],
[
[
"## MCA Analysis",
"_____no_output_____"
],
[
"### Remove unbalanced columns",
"_____no_output_____"
]
],
[
[
"features_df = product_cluster.iloc[:,3:-2]\nplt.figure(figsize=(16,20))\nfeatures = features_df.columns\nfor i,f in enumerate(features):\n counts = features_df.groupby([f])[f].count().to_dict()\n dist = list(counts.values())\n keys = list(counts.keys())\n chisq, p = chisquare(dist)\n plt.subplot(6,3,i+1)\n plt.title(\"%s\"%(f))\n plt.bar(range(len(keys)),dist)\n plt.xticks(range(len(keys)),keys,rotation=70)\n if len(keys)>10: plt.tick_params(axis='x', which='both', bottom=False, top=False, labelbottom=False)\nplt.subplots_adjust(wspace=0.5, hspace=0.5) \nplt.show()",
"_____no_output_____"
]
],
[
[
"### Apply MCA on Products",
"_____no_output_____"
]
],
[
[
"import prince\n\nunbalanced = [\"CANALE DISTRIB (Description)\",\"CLASSE COGE (Description)\",\"BIOLOGICO (Description)\"]\n\n\nfeatures_df = product_cluster.iloc[:,1:-2].drop(unbalanced,axis=1)\nfeatures_df = df_produit\nmca = prince.MCA(features_df)\nmca.plot_relationship_square()\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Apply MCA on Clients",
"_____no_output_____"
]
],
[
[
"import prince\nfeatures_df = client_df.astype(str).fillna(\"NA\")\nmca = prince.MCA(features_df)\nmca.plot_relationship_square()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Classification Tree",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split \nfrom sklearn.tree import DecisionTreeClassifier \nimport subprocess\nfrom sklearn.tree import export_graphviz\nfrom sklearn.preprocessing import OneHotEncoder,LabelBinarizer,LabelEncoder\n\n\ndef visualize_tree(tree, feature_names,class_names=None):\n \n with open(reports_path+\"dt.dot\", 'w') as f:\n \n export_graphviz(tree, out_file=f, feature_names=feature_names, filled=True, rounded=True, class_names=class_names )\n\n command = [\"C:\\\\Program Files (x86)\\\\Graphviz2.38\\\\bin\\\\dot.exe\", \"-Tpng\", reports_path+\"dt.dot\", \"-o\", \"dt.png\"]\n \n try:\n subprocess.check_call(command)\n except:\n exit(\"Could not run dot, ie graphviz, to \"\n \"produce visualization\")\n\n \ndrop = [\"Product Group key\",\"Centroid\",\"PRODOTTO (Description)\",\"PACKAGING (Description)\"]\nkeep = [\"FAM DETTAGLIATA (Description)\",\"FAM AGGREGATA (Description)\",\"MARCHIO (Description)\",\"GRUPPO MARCHIO (Description)\",\"SOTTO-TECNO (Description)\",\"CANALE DISTRIB (Description)\",\"CLASSE COGE (Description)\",\"FAM MARKETING (Description)\",\"Cluster\"]\n \n#data = product_cluster.drop(drop,axis=1)\ndata = product_cluster[keep]\n\n\n# cat_data = []\n# i=0\n# for label,col in data.iteritems():\n# cat_data.append(col.astype('category'))\n \n# df = pd.DataFrame(np.array(cat_data).T,columns = data.columns)\n\nlb = LabelBinarizer()\n\nX = pd.get_dummies(data.drop([\"Cluster\"],axis=1).iloc[:,:])\nprint(X.shape)\ndisplay(data.head())\nfeatures = X.columns\ny = lb.fit_transform(data.values[:,-1].astype(int).T)\ny = data.values[:,-1].astype(int)",
"(382, 125)\n"
],
[
"x_data = data.drop([\"Cluster\"],axis=1).iloc[:,:]\ny_data = data.values[:,-1].astype(int)\n\n\nfrom sklearn.feature_extraction import DictVectorizer\n\nX_dict = x_data.T.to_dict().values()\n\n\nvect = DictVectorizer(sparse=False)\nX_vector = vect.fit_transform(X_dict)\nprint(X_vector.shape)\ny = lb.fit_transform(y_data.T)\ny = y_data.T\nX = X_vector",
"(382, 125)\n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2) \n\nclassifier = DecisionTreeClassifier(criterion = \"gini\", max_depth=None, min_samples_leaf=1) \nclassifier.fit(X_train, y_train)\n\n\n# from sklearn.svm import SVC\n# classifier = SVC()\n# classifier.fit(X_train, y_train) \n\ny_pred = classifier.predict(X_test)",
"_____no_output_____"
]
],
[
[
"## Evaluation the algorithm",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report, confusion_matrix \nfrom sklearn.metrics import precision_recall_fscore_support as report\n#print(confusion_matrix(y_test, y_pred)) \nprecision,recall,fscore,support = report(y_test, y_pred,warn_for=())\nprint(precision.mean(),recall.mean(),fscore.mean(),support.sum())\n #print(classification_report(y_test, y_pred))\n\n# imp = np.array(classifier.feature_importances_)\n# imp_ft = features[np.argsort(imp)[::-1]]\n# print(imp_ft.values)",
"0.163285836074 0.153725490196 0.148187373144 77\n"
],
[
"visualize_tree(classifier, features,class_names=True)",
"_____no_output_____"
],
[
"from graphviz import Graph,Source\nfrom IPython.display import SVG\n\ngraph = Source(export_graphviz(classifier, out_file=None\n , feature_names=features, class_names=True\n , filled = True))\n\ndisplay(SVG(graph.pipe(format='svg')))",
"_____no_output_____"
],
[
"print(classifier.tree_)",
"<sklearn.tree._tree.Tree object at 0x000000000A407C60>\n"
]
]
] | [
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0b5265c177e4301d908994634695ce4e2006d79 | 8,971 | ipynb | Jupyter Notebook | practical_classes/Week_7/plot_decoding_csp_timefreq_practical_7.ipynb | victor-m-p/adv_cognitive_neuroscience | fdb2d4246fd1e934696c74090981431539868c3d | [
"MIT"
] | 2 | 2020-08-28T11:39:43.000Z | 2020-10-14T08:31:20.000Z | practical_classes/Week_7/plot_decoding_csp_timefreq_practical_7.ipynb | victor-m-p/adv_cognitive_neuroscience | fdb2d4246fd1e934696c74090981431539868c3d | [
"MIT"
] | null | null | null | practical_classes/Week_7/plot_decoding_csp_timefreq_practical_7.ipynb | victor-m-p/adv_cognitive_neuroscience | fdb2d4246fd1e934696c74090981431539868c3d | [
"MIT"
] | 5 | 2020-09-07T10:15:44.000Z | 2020-09-16T12:18:25.000Z | 33.349442 | 88 | 0.559692 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Decoding in time-frequency space using Common Spatial Patterns (CSP)\n\nThe time-frequency decomposition is estimated by iterating over raw data that\nhas been band-passed at different frequencies. This is used to compute a\ncovariance matrix over each epoch or a rolling time-window and extract the CSP\nfiltered signals. A linear discriminant classifier is then applied to these\nsignals.\n",
"_____no_output_____"
]
],
[
[
"# Authors: Laura Gwilliams <[email protected]>\n# Jean-Remi King <[email protected]>\n# Alex Barachant <[email protected]>\n# Alexandre Gramfort <[email protected]>\n#\n# License: BSD (3-clause)\n\n\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom mne import Epochs, create_info, events_from_annotations\nfrom mne.io import concatenate_raws, read_raw_edf\nfrom mne.datasets import eegbci\nfrom mne.decoding import CSP\nfrom mne.time_frequency import AverageTFR\n\nfrom sklearn.discriminant_analysis import LinearDiscriminantAnalysis\nfrom sklearn.model_selection import StratifiedKFold, cross_val_score\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.preprocessing import LabelEncoder",
"_____no_output_____"
]
],
[
[
"Set parameters and read data\n\n",
"_____no_output_____"
]
],
[
[
"event_id = dict(hands=2, feet=3) # motor imagery: hands vs feet\nsubject = 1\nruns = [6, 10, 14]\nraw_fnames = eegbci.load_data(subject, runs)\nraw = concatenate_raws([read_raw_edf(f) for f in raw_fnames])\n\n# Extract information from the raw file\nsfreq = raw.info['sfreq']\nevents, _ = events_from_annotations(raw, event_id=dict(T1=2, T2=3))\nraw.pick_types(meg=False, eeg=True, stim=False, eog=False, exclude='bads')\nraw.load_data()\n\n# Assemble the classifier using scikit-learn pipeline\nclf = make_pipeline(CSP(n_components=4, reg=None, log=True, norm_trace=False),\n LinearDiscriminantAnalysis())\nn_splits = 5 # how many folds to use for cross-validation\ncv = StratifiedKFold(n_splits=n_splits, shuffle=True, random_state=42)\n\n# Classification & time-frequency parameters\ntmin, tmax = -.200, 2.000\nn_cycles = 10. # how many complete cycles: used to define window size\nmin_freq = 5.\nmax_freq = 25.\nn_freqs = 8 # how many frequency bins to use\n\n# Assemble list of frequency range tuples\nfreqs = np.linspace(min_freq, max_freq, n_freqs) # assemble frequencies\nfreq_ranges = list(zip(freqs[:-1], freqs[1:])) # make freqs list of tuples\n\n# Infer window spacing from the max freq and number of cycles to avoid gaps\nwindow_spacing = (n_cycles / np.max(freqs) / 2.)\ncentered_w_times = np.arange(tmin, tmax, window_spacing)[1:]\nn_windows = len(centered_w_times)\n\n# Instantiate label encoder\nle = LabelEncoder()",
"_____no_output_____"
]
],
[
[
"Loop through frequencies, apply classifier and save scores\n\n",
"_____no_output_____"
]
],
[
[
"# init scores\nfreq_scores = np.zeros((n_freqs - 1,))\n\n# Loop through each frequency range of interest\nfor freq, (fmin, fmax) in enumerate(freq_ranges):\n\n # Infer window size based on the frequency being used\n w_size = n_cycles / ((fmax + fmin) / 2.) # in seconds\n\n # Apply band-pass filter to isolate the specified frequencies\n raw_filter = raw.copy().filter(fmin, fmax, n_jobs=1, fir_design='firwin',\n skip_by_annotation='edge')\n\n # Extract epochs from filtered data, padded by window size\n epochs = Epochs(raw_filter, events, event_id, tmin - w_size, tmax + w_size,\n proj=False, baseline=None, preload=True)\n epochs.drop_bad()\n y = le.fit_transform(epochs.events[:, 2])\n\n X = epochs.get_data()\n\n # Save mean scores over folds for each frequency and time window\n freq_scores[freq] = np.mean(cross_val_score(estimator=clf, X=X, y=y,\n scoring='roc_auc', cv=cv,\n n_jobs=1), axis=0)",
"_____no_output_____"
]
],
[
[
"Plot frequency results\n\n",
"_____no_output_____"
]
],
[
[
"plt.bar(freqs[:-1], freq_scores, width=np.diff(freqs)[0],\n align='edge', edgecolor='black')\nplt.xticks(freqs)\nplt.ylim([0, 1])\nplt.axhline(len(epochs['feet']) / len(epochs), color='k', linestyle='--',\n label='chance level')\nplt.legend()\nplt.xlabel('Frequency (Hz)')\nplt.ylabel('Decoding Scores')\nplt.title('Frequency Decoding Scores')",
"_____no_output_____"
]
],
[
[
"Loop through frequencies and time, apply classifier and save scores\n\n",
"_____no_output_____"
]
],
[
[
"# init scores\ntf_scores = np.zeros((n_freqs - 1, n_windows))\n\n# Loop through each frequency range of interest\nfor freq, (fmin, fmax) in enumerate(freq_ranges):\n\n # Infer window size based on the frequency being used\n w_size = n_cycles / ((fmax + fmin) / 2.) # in seconds\n\n # Apply band-pass filter to isolate the specified frequencies\n raw_filter = raw.copy().filter(fmin, fmax, n_jobs=1, fir_design='firwin',\n skip_by_annotation='edge')\n\n # Extract epochs from filtered data, padded by window size\n epochs = Epochs(raw_filter, events, event_id, tmin - w_size, tmax + w_size,\n proj=False, baseline=None, preload=True)\n epochs.drop_bad()\n y = le.fit_transform(epochs.events[:, 2])\n\n # Roll covariance, csp and lda over time\n for t, w_time in enumerate(centered_w_times):\n\n # Center the min and max of the window\n w_tmin = w_time - w_size / 2.\n w_tmax = w_time + w_size / 2.\n\n # Crop data into time-window of interest\n X = epochs.copy().crop(w_tmin, w_tmax).get_data()\n\n # Save mean scores over folds for each frequency and time window\n tf_scores[freq, t] = np.mean(cross_val_score(estimator=clf, X=X, y=y,\n scoring='roc_auc', cv=cv,\n n_jobs=1), axis=0)",
"_____no_output_____"
]
],
[
[
"Plot time-frequency results\n\n",
"_____no_output_____"
]
],
[
[
"# Set up time frequency object\nav_tfr = AverageTFR(create_info(['freq'], sfreq), tf_scores[np.newaxis, :],\n centered_w_times, freqs[1:], 1)\n\nchance = np.mean(y) # set chance level to white in the plot\nav_tfr.plot([0], vmin=chance, title=\"Time-Frequency Decoding Scores\",\n cmap=plt.cm.Reds)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b52ca75176f94b99304884170fc3ca1a4234bf | 16,665 | ipynb | Jupyter Notebook | week0_03_logistic/week03_extra_image_classifier.ipynb | GendalfSeriyy/ml-mipt | 647a482baba57d8b920392ed534c8179194dfea7 | [
"MIT"
] | 6 | 2021-11-17T18:34:34.000Z | 2022-01-18T18:29:07.000Z | week0_03_logistic/week03_extra_image_classifier.ipynb | GendalfSeriyy/ml-mipt | 647a482baba57d8b920392ed534c8179194dfea7 | [
"MIT"
] | null | null | null | week0_03_logistic/week03_extra_image_classifier.ipynb | GendalfSeriyy/ml-mipt | 647a482baba57d8b920392ed534c8179194dfea7 | [
"MIT"
] | 2 | 2020-09-30T21:22:47.000Z | 2021-01-05T14:44:01.000Z | 30.918367 | 475 | 0.589619 | [
[
[
"## week03: Логистическая регрессия и анализ изображений\n\n\nВ этом ноутбуке предлагается построить классификатор изображений на основе логистической регрессии. \n\n*Забегая вперед, мы попробуем решить задачу классификации изображений используя лишь простые методы. В третьей части нашего курса мы вернемся к этой задаче.*\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport h5py\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## 1. Постановка задачи ##\n\n\n**Задача**: Есть датасет [прямая ссылка](https://drive.google.com/file/d/15tOimf2QYWsMtPJXTUCwgZaOTF8Nxcsm/view?usp=sharing) (\"catvnoncat.h5\") состоящий из:\n - обучающей выборки из m_train изображений, помеченных \"cat\" (y=1) или \"non-cat\" (y=0)\n - тестовой выборки m_test изображений, помеченных \"cat\" или \"non-cat\"\n - каждое цветное изображение имеет размер (src_size, src_size, 3), где 3 - число каналов (RGB).\n Таким образом, каждый слой - квадрат размера src_size x src_size$.\n\nДавайте построим простой алгоритм классификации изображений на классы \"cat\"/\"non-cat\".\n\nАвтоматическая загрузка доступна ниже.",
"_____no_output_____"
],
[
"<img src=\"img/LogReg_kiank.png\" style=\"width:650px;height:400px;\">\n\n**Recap**:\n\nДля каждого примера $x^{(i)}$:\n$$z^{(i)} = w^T x^{(i)} + b \\tag{1}$$\n$$\\hat{y}^{(i)} = a^{(i)} = sigmoid(z^{(i)})\\tag{2}$$ \n$$ \\mathcal{L}(a^{(i)}, y^{(i)}) = - y^{(i)} \\log(a^{(i)}) - (1-y^{(i)} ) \\log(1-a^{(i)})\\tag{3}$$\n\nФункция потерь:\n$$ J = \\frac{1}{m} \\sum_{i=1}^m \\mathcal{L}(a^{(i)}, y^{(i)})\\tag{6}$$",
"_____no_output_____"
]
],
[
[
"# Uncomment this cell to download the data\n\n# !wget \"https://downloader.disk.yandex.ru/disk/7ef1d1e30e23740a4a30799a825319154815ddc85bf689542add0a3d11ccb91c/5d7fdcb0/3dcxK38Q0fG3ui0g2gMZgKkLls8ULwVpoYNkWpBm9d24EceJ6mIoH5l3_wKkFv3PfZ0WMGYjfJULynuJkuGaug%3D%3D?uid=76549735&filename=data.zip&disposition=attachment&hash=&limit=0&content_type=application%2Fzip&owner_uid=76549735&fsize=2815580&hid=084389255415f71a92d0f1024ab741d4&media_type=compressed&tknv=v2&etag=2b348ac8eca72d223108e36b2a671210\" -O data.zip\n# !unzip data.zip",
"_____no_output_____"
]
],
[
[
"### 1.1 Загрузка данных и визуализация ###",
"_____no_output_____"
]
],
[
[
"def load_dataset():\n train_data = h5py.File(\"data/train_catvnoncat.h5\", \"r\")\n train_set_x_orig = np.array(train_data[\"train_set_x\"][:]) # признаки\n train_set_y_orig = np.array(train_data[\"train_set_y\"][:]) # метки классов\n\n test_data = h5py.File(\"data/test_catvnoncat.h5\", \"r\")\n test_set_x_orig = np.array(test_data[\"test_set_x\"][:]) # признаки\n test_set_y_orig = np.array(test_data[\"test_set_y\"][:]) # метки классов\n\n classes = np.array(test_data[\"list_classes\"][:]) # the list of classes\n classes = np.array(list(map(lambda x: x.decode('utf-8'), classes)))\n \n train_set_y = train_set_y_orig.reshape(train_set_y_orig.shape[0])\n test_set_y = test_set_y_orig.reshape(test_set_y_orig.shape[0])\n return train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes",
"_____no_output_____"
],
[
"train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes = load_dataset()",
"_____no_output_____"
]
],
[
[
"Цветные изображения в формате RGB представлены в виде трёхмерных numpy.array.\n\nПорядок измерений $H \\times W \\times C$: $H$ - высота, $W$ - ширина и $C$ - число каналов.\n\nЗначение каждого пиксела находится в интервале $[0;255]$.",
"_____no_output_____"
]
],
[
[
"from ipywidgets import interact, interactive, fixed, interact_manual\nimport ipywidgets as widgets\n\ndef show_image_interact(i=0):\n f, ax = plt.subplots(1,4, figsize=(15,20), sharey=True)\n \n ax[0].imshow(train_set_x_orig[i])\n ax[0].set_title('RGB image')\n ax[1].imshow(train_set_x_orig[i][:,:,0], cmap='gray')\n ax[1].set_title('R channel')\n ax[2].imshow(train_set_x_orig[i][:,:,1], cmap='gray')\n ax[2].set_title('G channel')\n ax[3].imshow(train_set_x_orig[i][:,:,2], cmap='gray')\n ax[3].set_title('B channel')\n \n print(\"y = {} belongs to '{}' class.\".format(str(train_set_y[i]),classes[np.squeeze(train_set_y[i])]))\n\ninteract(show_image_interact,\n i=widgets.IntSlider(min=0, max=len(train_set_y)-1, step=1))",
"_____no_output_____"
]
],
[
[
"При работе с данными полезно будет сохранить размерности входных изображений для дальнейшей обработки.",
"_____no_output_____"
]
],
[
[
"m_train = train_set_x_orig.shape[0]\nm_test = test_set_x_orig.shape[0]\nsrc_size = train_set_x_orig.shape[1]\n\nprint (\"Размер обучающей выборки: m_train = \" + str(m_train))\nprint (\"Размер тестовой выборки: m_test = \" + str(m_test))\nprint (\"Ширина/Высота каждого изображения: src_size = \" + str(src_size))\nprint (\"Размерны трёхмерной матрицы для каждого изображения: (\" + str(src_size) + \", \" + str(src_size) + \", 3)\")\nprint (\"Размерность train_set_x: \" + str(train_set_x_orig.shape))\nprint (\"Размерность train_set_y: \" + str(train_set_y.shape))\nprint (\"Размерность test_set_x: \" + str(test_set_x_orig.shape))\nprint (\"Размерность test_set_y: \" + str(test_set_y.shape))",
"_____no_output_____"
]
],
[
[
"## 2. Предварительная обработка",
"_____no_output_____"
],
[
"Преобразуем входные изображения размера (num_px, num_px, 3) в вектор признаков размера (num_px $*$ num_px $*$ 3, 1), чтобы сформировать матрицы объект-признак в виде numpy-array для обучающей и тестовой выборок.\n\nКаждой строке матрицы объект-признак соответствует входное развёрнутое в вектор-строку изображение.\n\nПомимо этого, для предварительной обработки (препроцессинга) изображений применяют центрирование значений: из значения каждого пиксела вычитается среднее и делят полученное значение на среднеквадратичное отклонение значений пикселей всего изображения.\n\nОднако, на практике обычно просто делят значения пикселей на 255 (максимальное значение пикселя).\n\nОформим эти шаги в функцию предварительной обработки",
"_____no_output_____"
]
],
[
[
"def image_preprocessing_simple(data):\n assert type(data) == np.ndarray\n assert data.ndim == 4\n \n n,h,w,c = data.shape\n data_vectorized = <ваш код>\n data_normalized = <ваш код>\n \n return data_normalized",
"_____no_output_____"
],
[
"# Изменить размеры входных данных\n\ntrain_set_x_vectorized = image_preprocessing_simple(train_set_x_orig)\ntest_set_x_vectorized = image_preprocessing_simple(test_set_x_orig)\n\nprint('Train set:')\nprint(\"Размеры train_set_x_vectorized: {}\".format(str(train_set_x_vectorized.shape)))\nprint(\"Размеры train_set_y: {}\".format(str(train_set_y.shape)))\nprint(\"Размеры классов 'cat'/'non-cat': {} / {}\".format(sum(train_set_y==1), sum(train_set_y==0)))\nprint('Test set:')\nprint(\"Размеры test_set_x_vectorized: {}\".format(str(test_set_x_vectorized.shape)))\nprint(\"Размеры test_set_y: {}\".format(str(test_set_y.shape)))\nprint(\"Размеры классов 'cat'/'non-cat': {} / {}\".format(sum(test_set_y==1), sum(test_set_y==0)))",
"_____no_output_____"
]
],
[
[
"## 3. Классификация",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import GridSearchCV\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"**Вопрос**: Какую метрику качества стоит использовать?",
"_____no_output_____"
],
[
"### 3.1 Построение модели",
"_____no_output_____"
],
[
"Построим модель с параметрами по умолчанию и посмотрим, как хорошо она справится с задачей.",
"_____no_output_____"
]
],
[
[
"clf = # <ваш код>\n\nscore = <ваш код> \nprint('Точность для простой модели с параметрами по умолчанию: {:.4f}'.format(score))",
"_____no_output_____"
],
[
"# from sklearn.metrics import f1_score\n\ny_predicted = clf.predict(test_set_x_vectorized)\ncorrect_score = <ваш код>\nprint('<Имя метрики> для простой модели: {:.4f}'.format(correct_score))",
"_____no_output_____"
]
],
[
[
"Попробуем подобрать параметры регуляризации в надежде, что это повысит точность предсказаний.",
"_____no_output_____"
]
],
[
[
"<ваш код>",
"_____no_output_____"
],
[
"print('Оптимальные параметры: {}'.format(<ваш код>))\nprint('Наилучшее значение метрики качества: {}'.format(<ваш код>))",
"_____no_output_____"
]
],
[
[
"Обучим модель с оптимальными параметрами на всей обучающей выборке и посмотрим на метрики качества:",
"_____no_output_____"
]
],
[
[
"best_clf = <ваш код>\nbest_clf.fit(train_set_x_vectorized, train_set_y)\n\ny_predicted = best_clf.predict(test_set_x_vectorized)\nmetric_score = <ваш код>(y_predicted, test_set_y)\nprint('Optimal model hyperparameters accuracy score: {:.4f}'.format(metric_score))",
"_____no_output_____"
]
],
[
[
"### 3.2 Анализ ошибок",
"_____no_output_____"
]
],
[
[
"is_outlier = (y_predicted != test_set_y)\ntest_outliers_x, test_outliers_y, predicted_y = test_set_x_orig[is_outlier], test_set_y[is_outlier], y_predicted[is_outlier]",
"_____no_output_____"
],
[
"def show_image_outliers(i=0):\n f = plt.figure(figsize=(5,5))\n plt.imshow(test_outliers_x[i])\n plt.title('RGB image')\n \n fmt_string = \"Sample belongs to '{}' class, but '{}' is predicted'\"\n print(fmt_string.format(classes[test_outliers_y[i]], classes[predicted_y[i]]))\n\ninteract(show_image_outliers,\n i=widgets.IntSlider(min=0, max=len(test_outliers_y)-1, step=1))\n",
"_____no_output_____"
]
],
[
[
"**Вопрос**: Как по-вашему можно повысить точность? Каким недостатком обладает данный подход к классификации?",
"_____no_output_____"
],
[
"### 3.3 Модель с аугментациями",
"_____no_output_____"
],
[
"Как можно увеличить количество данных для обучения?\n\nСформировать новые примеры из уже имеющихся!\n\nНапример, можно пополнить class 'cat' обучающей выборки [зеркально отображёнными](https://docs.scipy.org/doc/numpy/reference/generated/numpy.fliplr.html) изображениями котов.",
"_____no_output_____"
]
],
[
[
"def augment_sample(src, label):\n <ваш код>\n\ndef image_preprocessing_augment(data, labels):\n assert type(data) == np.ndarray\n assert data.ndim == 4\n \n ## ВАШ КОД ##\n \n \n data_augmented = \n labels_augmented = \n ## ВАШ КОД ЗАКАНЧИВАЕТСЯ ЗДЕСЬ ##\n \n n,h,w,c = data_augmented.shape\n data_vectorized = data_augmented.reshape(n, -1) # <ваш код>\n data_normalized = data_vectorized / 255\n \n return data_normalized, labels_augmented",
"_____no_output_____"
],
[
"train_set_x_augmented, train_set_y_augmented = image_preprocessing_augment(train_set_x_orig, train_set_y)",
"_____no_output_____"
],
[
"clf = LogisticRegression(solver='liblinear')\nclf.fit(train_set_x_augmented, train_set_y_augmented)\ny_pred = clf.predict(test_set_x_vectorized)\nprint('F-мера для модели с аугментациями: {:.4f}'.format(f1_score(y_pred, test_set_y)))",
"_____no_output_____"
]
],
[
[
"## 4. Проверьте работу классификатора на своей картинке",
"_____no_output_____"
],
[
"Библиотека [OpenCV](https://opencv.org) для работы с изображениями для [python](https://pypi.org/project/opencv-python/):\n\n`pip install opencv-python`\n\nВместе с contrib-модулями:\n\n`pip install opencv-contrib-python`\n",
"_____no_output_____"
]
],
[
[
"import cv2\n\n# Путь к картинке на вашем ПК\nfname = \"cat-non-cat.jpg\"\n# Считываем картинку через scipy\nsrc = cv2.cvtColor(cv2.imread((fname)), cv2.COLOR_BGR2RGB)\nsrc_resized = cv2.resize(src, (src_size,src_size), interpolation=cv2.INTER_LINEAR).reshape(1, src_size*src_size*3)\nmy_image_predict = clf.predict(src_resized)[0]\n\nplt.imshow(src)\nprint(\"Алгоритм говорит, что это '{}': {}\".format(my_image_predict, classes[my_image_predict]))",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
d0b52e619dec46590fa50affde0de56a1ad2cdb6 | 12,408 | ipynb | Jupyter Notebook | Spark_MLlib/NaturalLP/EmailSpan.ipynb | Dev566/SelfLearning-PySpark | c967f8c34c8d5c840ebd185afb3b80f64f694993 | [
"Apache-2.0"
] | null | null | null | Spark_MLlib/NaturalLP/EmailSpan.ipynb | Dev566/SelfLearning-PySpark | c967f8c34c8d5c840ebd185afb3b80f64f694993 | [
"Apache-2.0"
] | null | null | null | Spark_MLlib/NaturalLP/EmailSpan.ipynb | Dev566/SelfLearning-PySpark | c967f8c34c8d5c840ebd185afb3b80f64f694993 | [
"Apache-2.0"
] | 1 | 2019-11-09T00:05:34.000Z | 2019-11-09T00:05:34.000Z | 23.063197 | 115 | 0.458172 | [
[
[
"import findspark\nfindspark.init('/home/ubuntu/spark-2.1.1-bin-hadoop2.7')\nimport pyspark\nfrom pyspark.sql import SparkSession\nspark = SparkSession.builder.appName('EmailSpan').getOrCreate()",
"_____no_output_____"
],
[
"from pyspark.ml.feature import Tokenizer,RegexTokenizer,IDF,CountVectorizer,StringIndexer,StopWordsRemover\nfrom pyspark.sql.functions import col,udf,length\nfrom pyspark.sql.types import IntegerType\n",
"_____no_output_____"
],
[
"data=spark.read.csv('SMSSpamCollection',inferSchema=True,sep='\\t')",
"_____no_output_____"
],
[
"data.show()",
"+----+--------------------+\n| _c0| _c1|\n+----+--------------------+\n| ham|Go until jurong p...|\n| ham|Ok lar... Joking ...|\n|spam|Free entry in 2 a...|\n| ham|U dun say so earl...|\n| ham|Nah I don't think...|\n|spam|FreeMsg Hey there...|\n| ham|Even my brother i...|\n| ham|As per your reque...|\n|spam|WINNER!! As a val...|\n|spam|Had your mobile 1...|\n| ham|I'm gonna be home...|\n|spam|SIX chances to wi...|\n|spam|URGENT! You have ...|\n| ham|I've been searchi...|\n| ham|I HAVE A DATE ON ...|\n|spam|XXXMobileMovieClu...|\n| ham|Oh k...i'm watchi...|\n| ham|Eh u remember how...|\n| ham|Fine if thats th...|\n|spam|England v Macedon...|\n+----+--------------------+\nonly showing top 20 rows\n\n"
],
[
"data = data.withColumnRenamed('_c0','class').withColumnRenamed('_c1','text')",
"_____no_output_____"
],
[
"data.show()",
"+-----+--------------------+\n|class| text|\n+-----+--------------------+\n| ham|Go until jurong p...|\n| ham|Ok lar... Joking ...|\n| spam|Free entry in 2 a...|\n| ham|U dun say so earl...|\n| ham|Nah I don't think...|\n| spam|FreeMsg Hey there...|\n| ham|Even my brother i...|\n| ham|As per your reque...|\n| spam|WINNER!! As a val...|\n| spam|Had your mobile 1...|\n| ham|I'm gonna be home...|\n| spam|SIX chances to wi...|\n| spam|URGENT! You have ...|\n| ham|I've been searchi...|\n| ham|I HAVE A DATE ON ...|\n| spam|XXXMobileMovieClu...|\n| ham|Oh k...i'm watchi...|\n| ham|Eh u remember how...|\n| ham|Fine if thats th...|\n| spam|England v Macedon...|\n+-----+--------------------+\nonly showing top 20 rows\n\n"
],
[
"data = data.withColumn('lenght',length(data['text']))",
"_____no_output_____"
],
[
"data.show()",
"+-----+--------------------+------+\n|class| text|lenght|\n+-----+--------------------+------+\n| ham|Go until jurong p...| 111|\n| ham|Ok lar... Joking ...| 29|\n| spam|Free entry in 2 a...| 155|\n| ham|U dun say so earl...| 49|\n| ham|Nah I don't think...| 61|\n| spam|FreeMsg Hey there...| 147|\n| ham|Even my brother i...| 77|\n| ham|As per your reque...| 160|\n| spam|WINNER!! As a val...| 157|\n| spam|Had your mobile 1...| 154|\n| ham|I'm gonna be home...| 109|\n| spam|SIX chances to wi...| 136|\n| spam|URGENT! You have ...| 155|\n| ham|I've been searchi...| 196|\n| ham|I HAVE A DATE ON ...| 35|\n| spam|XXXMobileMovieClu...| 149|\n| ham|Oh k...i'm watchi...| 26|\n| ham|Eh u remember how...| 81|\n| ham|Fine if thats th...| 56|\n| spam|England v Macedon...| 155|\n+-----+--------------------+------+\nonly showing top 20 rows\n\n"
],
[
"data.groupBy('class').mean().show()",
"+-----+-----------------+\n|class| avg(lenght)|\n+-----+-----------------+\n| ham|71.45431945307645|\n| spam|138.6706827309237|\n+-----+-----------------+\n\n"
],
[
"indexer = StringIndexer(inputCol='class',outputCol='label')",
"_____no_output_____"
],
[
"#index_model = indexer.fit(data)",
"_____no_output_____"
],
[
"#index_data = index_model.transform(data)",
"_____no_output_____"
],
[
"tokenizer = Tokenizer(inputCol='text',outputCol='tok_word')",
"_____no_output_____"
],
[
"#tokenized_data = tokenizer.transform(index_data)",
"_____no_output_____"
],
[
"stop_word = StopWordsRemover(inputCol='tok_word',outputCol='stop_token')",
"_____no_output_____"
],
[
"#stop_data = stop_word.transform(tokenized_data)",
"_____no_output_____"
],
[
"count_vec = CountVectorizer(inputCol='stop_token',outputCol='c_vec')",
"_____no_output_____"
],
[
"idf = IDF(inputCol='c_vec',outputCol='tf_idf')",
"_____no_output_____"
],
[
"from pyspark.ml.feature import VectorAssembler",
"_____no_output_____"
],
[
"clean_up = VectorAssembler(inputCols=['tf_idf','lenght'],outputCol='features')",
"_____no_output_____"
],
[
"from pyspark.ml.classification import NaiveBayes",
"_____no_output_____"
],
[
"nb = NaiveBayes()",
"_____no_output_____"
],
[
"from pyspark.ml import Pipeline",
"_____no_output_____"
],
[
"data_prep_pipe = Pipeline(stages=[indexer,tokenizer,stop_word,count_vec,idf,clean_up])",
"_____no_output_____"
],
[
"cleaner = data_prep_pipe.fit(data)",
"_____no_output_____"
],
[
"clean_data = cleaner.transform(data)",
"_____no_output_____"
],
[
"clean_data.printSchema()",
"root\n |-- class: string (nullable = true)\n |-- text: string (nullable = true)\n |-- lenght: integer (nullable = true)\n |-- label: double (nullable = true)\n |-- tok_word: array (nullable = true)\n | |-- element: string (containsNull = true)\n |-- stop_token: array (nullable = true)\n | |-- element: string (containsNull = true)\n |-- c_vec: vector (nullable = true)\n |-- tf_idf: vector (nullable = true)\n |-- features: vector (nullable = true)\n\n"
],
[
"final_data = clean_data.select('label','features')",
"_____no_output_____"
],
[
"training_data,test_data = final_data.randomSplit([0.7,0.3])",
"_____no_output_____"
],
[
"spam_model = nb.fit(training_data)",
"_____no_output_____"
],
[
"test_result= spam_model.transform(test_data)",
"_____no_output_____"
],
[
"test_result.select('label','prediction').show()",
"+-----+----------+\n|label|prediction|\n+-----+----------+\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 1.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n| 0.0| 0.0|\n+-----+----------+\nonly showing top 20 rows\n\n"
],
[
"from pyspark.ml.evaluation import MulticlassClassificationEvaluator",
"_____no_output_____"
],
[
"m_eval = MulticlassClassificationEvaluator()",
"_____no_output_____"
],
[
"m_eval.evaluate(test_result)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b53866095ef8b1b6f171f33c955b61caee008d | 33,766 | ipynb | Jupyter Notebook | preprocessing/tweet_features.ipynb | RecKIE7/recsys2021-twitter | 72f1cd1c0db84110b682684d588b24665860683d | [
"Apache-2.0"
] | null | null | null | preprocessing/tweet_features.ipynb | RecKIE7/recsys2021-twitter | 72f1cd1c0db84110b682684d588b24665860683d | [
"Apache-2.0"
] | null | null | null | preprocessing/tweet_features.ipynb | RecKIE7/recsys2021-twitter | 72f1cd1c0db84110b682684d588b24665860683d | [
"Apache-2.0"
] | null | null | null | 63.350844 | 8,300 | 0.461914 | [
[
[
"# Extracted from text token\ntweet_feature_mentions: list of ints (or None):\n- Mentions extracted from the tweet.\n\ntweet_feature_number_of_mentions: int:\n- Number of mentions in the tweet.\n\ntweet_feature_token_length: int:\n- Number of BERT tokens in the tweet.\n\ntweet_feature_token_length_unique: int:\n- Number of unique bert tokens in the tweet.\n\ntweet_feature_text_token_decoded: list of str:\n- Decoded BERT tokens.\n\ntweet_feature_text_topic_word_count_adult_content: int:\n- Number of 'adult content' words.\n\ntweet_feature_text_topic_word_count_kpop: int:\n- Number of 'kpop' words.\n\ntweet_feature_text_topic_word_count_covid: int:\n- Number of 'covid' words.\n\ntweet_feature_text_topic_word_count_sport: int:\n- Number of 'sport' words.",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('..')\n\nimport core.config as conf\nfrom utils.preprocessing import *\nimport numpy as np\nfrom tqdm import tqdm\nfrom datetime import datetime \nimport matplotlib.pyplot as plt\n\nimport tensorflow as tf\nfrom transformers import *\nfrom tensorflow.keras.preprocessing.sequence import pad_sequences\nfrom tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint\n",
"_____no_output_____"
],
[
"tqdm.pandas()",
"_____no_output_____"
],
[
"#random seed \ntf.random.set_seed(1234)\nnp.random.seed(1234)",
"_____no_output_____"
],
[
"tokenizer = BertTokenizer.from_pretrained(\"bert-base-multilingual-cased\", cache_dir='bert_ckpt', do_lower_case=False)",
"_____no_output_____"
]
],
[
[
"## Load data",
"_____no_output_____"
]
],
[
[
"data_path = '/hdd/twitter/dataset_mini/train'\ndf = read_data(data_path)",
"_____no_output_____"
],
[
"text_tokens = df['text_tokens']",
"_____no_output_____"
],
[
"text_tokens",
"_____no_output_____"
],
[
"df['len_text_tokens'] = df['text_tokens'].apply(lambda x: len(x.split('\\t')))",
"_____no_output_____"
],
[
"df['decoded_text_tokens'] = df['text_tokens'].progress_apply(lambda x: tokenizer.decode(x.split('\\t'), skip_special_tokens=True))",
"100%|██████████| 4338906/4338906 [43:49<00:00, 1649.95it/s]\n"
],
[
"# x = '101\\t56898\\t137'\n# tokenizer.decode(x.split('\\t'), skip_special_tokens=True)",
"_____no_output_____"
],
[
"df['cnt_mention'] = df['text_tokens'].progress_apply(lambda x: (x.split('\\t').count('137')))",
"100%|██████████| 4338906/4338906 [00:08<00:00, 506917.75it/s]\n"
],
[
"df['len_text_tokens_unique'] = df['text_tokens'].progress_apply(lambda x: len(list(set(x.split('\\t')))))",
"100%|██████████| 4338906/4338906 [00:19<00:00, 217804.30it/s]\n"
],
[
"df.head()",
"_____no_output_____"
],
[
"tokenizer.encode('adult content')",
"_____no_output_____"
],
[
"tokenizer.decode([101, 11170, 32194, 102])",
"_____no_output_____"
],
[
"df['wc_sport'] = df['text_tokens'].progress_apply(lambda x: (x.split('\\t').count('17925')))",
"100%|██████████| 4338906/4338906 [00:08<00:00, 483630.57it/s]\n"
],
[
"df",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b53e97667737c3cf3b11d94f4f453961b92ee4 | 8,667 | ipynb | Jupyter Notebook | ipnb/Parallel ScalableKMeansPP.ipynb | thilinamb/k-means-parallel | 05331b48f2b354412eb0e8874a38d5df2bfe15d6 | [
"Apache-2.0"
] | 7 | 2015-12-04T08:27:44.000Z | 2020-11-22T07:24:31.000Z | ipnb/Parallel ScalableKMeansPP.ipynb | thilinamb/k-means-parallel | 05331b48f2b354412eb0e8874a38d5df2bfe15d6 | [
"Apache-2.0"
] | null | null | null | ipnb/Parallel ScalableKMeansPP.ipynb | thilinamb/k-means-parallel | 05331b48f2b354412eb0e8874a38d5df2bfe15d6 | [
"Apache-2.0"
] | 5 | 2015-04-25T21:01:53.000Z | 2019-05-30T17:54:26.000Z | 38.52 | 132 | 0.547133 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b53f9414a7e7b45245d48fa2b6c9d6877019c4 | 6,099 | ipynb | Jupyter Notebook | Python3/3_Using_results.ipynb | fabiolib/rest-api-jupyter-course | c3fd4a36cbfd86ce1571d1056a78ffef93395df6 | [
"Apache-2.0"
] | null | null | null | Python3/3_Using_results.ipynb | fabiolib/rest-api-jupyter-course | c3fd4a36cbfd86ce1571d1056a78ffef93395df6 | [
"Apache-2.0"
] | null | null | null | Python3/3_Using_results.ipynb | fabiolib/rest-api-jupyter-course | c3fd4a36cbfd86ce1571d1056a78ffef93395df6 | [
"Apache-2.0"
] | null | null | null | 26.402597 | 333 | 0.520085 | [
[
[
"# Using results\n\nSince json is a dictionary, you can pull out a single datapoint using the key.\n\n```\n{\n \"source\": \"ensembl_havana\",\n \"object_type\": \"Gene\",\n \"logic_name\": \"ensembl_havana_gene\",\n \"version\": 12,\n \"species\": \"homo_sapiens\",\n \"description\": \"B-Raf proto-oncogene, serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1097]\",\n \"display_name\": \"BRAF\",\n \"assembly_name\": \"GRCh38\",\n \"biotype\": \"protein_coding\",\n \"end\": 140924764,\n \"seq_region_name\": \"7\",\n \"db_type\": \"core\",\n \"strand\": -1,\n \"id\": \"ENSG00000157764\",\n \"start\": 140719327\n}\n```\n\nWe can add this to our previous script:",
"_____no_output_____"
]
],
[
[
"import requests, json\nfrom pprint import pprint\n\ndef fetch_endpoint(server, request, content_type):\n\n r = requests.get(server+request, headers={ \"Accept\" : content_type})\n\n if not r.ok:\n r.raise_for_status()\n sys.exit()\n\n if content_type == 'application/json':\n return r.json()\n else:\n return r.text\n\n\nserver = \"http://rest.ensembl.org/\"\next = \"lookup/id/ENSG00000157764?\"\ncon = \"application/json\"\nget_gene = fetch_endpoint(server, ext, con)\n\nsymbol = get_gene['display_name']\nprint (symbol)",
"BRAF\n"
]
],
[
[
"## Exercises 3\n\n1\\. Write a script to lookup the gene called *ESPN* in human and print the stable ID of this gene.",
"_____no_output_____"
]
],
[
[
"# Exercise 3.1\n\n#!/usr/bin/env python\n\n# Get modules needed for script\nimport sys, requests, json\nfrom pprint import pprint\n\ndef fetch_endpoint(server, request, content_type):\n\n r = requests.get(server+request, headers={ \"Accept\" : content_type})\n\n if not r.ok:\n r.raise_for_status()\n sys.exit()\n\n if content_type == 'application/json':\n return r.json()\n else:\n return r.text\n\n# define the gene name\ngene_name = \"ESPN\"\n\n# define the general URL parameters\nserver = \"http://rest.ensembl.org/\"\n\n# define REST query to get the gene ID from the gene name\next_get_lookup = \"lookup/symbol/homo_sapiens/\" + gene_name + \"?\"\n\n# define the content type\ncon = \"application/json\"\n\n# submit the query\nget_lookup = fetch_endpoint(server, ext_get_lookup, con)\n\n#pprint(get_lookup)\npprint(get_lookup['id'])",
"'ENSG00000187017'\n"
]
],
[
[
"2\\. Get all variants that are associated with the phenotype 'Coffee consumption'. For each variant print\n\n a. the p-value for the association\n \n b. the PMID for the publication which describes the association between that variant and ‘Coffee consumption’\n \n c. the risk allele and the associated gene.",
"_____no_output_____"
]
],
[
[
"# Exercise 3.2\n\npprint(get_lookup)",
"{'assembly_name': 'GRCh38',\n 'biotype': 'protein_coding',\n 'db_type': 'core',\n 'description': 'espin [Source:HGNC Symbol;Acc:HGNC:13281]',\n 'display_name': 'ESPN',\n 'end': 6461367,\n 'id': 'ENSG00000187017',\n 'logic_name': 'ensembl_havana_gene_homo_sapiens',\n 'object_type': 'Gene',\n 'seq_region_name': '1',\n 'source': 'ensembl_havana',\n 'species': 'homo_sapiens',\n 'start': 6424776,\n 'strand': 1,\n 'version': 17}\n"
]
],
[
[
"3\\. Get the mouse homologue of the human BRCA2 and print the ID and sequence of both.\n\nNote that the JSON for the endpoint you need is several layers deep, containing nested lists (appear as square brackets [ ] in the JSON) and key value sets (dictionary; appear as curly brackets { } in the JSON). Pretty print (pprint) comes in very useful here for the intermediate stage when you're trying to work out the json.",
"_____no_output_____"
]
],
[
[
"# Exercise 3.3",
"_____no_output_____"
]
],
[
[
"[Next page: Exercises 3 – answers](3_Using_results_answers.ipynb)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b540fc75618bf186167764956b135133766509 | 131 | ipynb | Jupyter Notebook | notebooks/App.ipynb | BatoolMM/PlantAI | 8626715928e9c50e6acb774e1c3c7a0417f6175c | [
"MIT"
] | 5 | 2020-10-05T06:18:15.000Z | 2021-09-22T15:48:19.000Z | notebooks/App.ipynb | BatoolMM/PlantAI | 8626715928e9c50e6acb774e1c3c7a0417f6175c | [
"MIT"
] | null | null | null | notebooks/App.ipynb | BatoolMM/PlantAI | 8626715928e9c50e6acb774e1c3c7a0417f6175c | [
"MIT"
] | null | null | null | 32.75 | 75 | 0.885496 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b554cd1abe8da438a3da1207fb2e2bfa78afe0 | 4,680 | ipynb | Jupyter Notebook | Face Detection ML/Face Detection.ipynb | Aastha3348/Face-Detection-using-Machine-Learning | f4287a8e7493c10f3c8a229b789ea10aa6d1a889 | [
"Apache-2.0"
] | null | null | null | Face Detection ML/Face Detection.ipynb | Aastha3348/Face-Detection-using-Machine-Learning | f4287a8e7493c10f3c8a229b789ea10aa6d1a889 | [
"Apache-2.0"
] | null | null | null | Face Detection ML/Face Detection.ipynb | Aastha3348/Face-Detection-using-Machine-Learning | f4287a8e7493c10f3c8a229b789ea10aa6d1a889 | [
"Apache-2.0"
] | null | null | null | 21.869159 | 82 | 0.386538 | [
[
[
"import cv2",
"_____no_output_____"
],
[
"faces = cv2.CascadeClassifier(\"haarcascade_frontalface_alt.xml\")",
"_____no_output_____"
],
[
"img = cv2.imread(\"Emma Stone.jpg\")",
"_____no_output_____"
],
[
"img",
"_____no_output_____"
],
[
"img_gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)",
"_____no_output_____"
],
[
"img_gray",
"_____no_output_____"
],
[
"detections = faces.detectMultiScale(img_gray,scaleFactor=1.1,minNeighbors=6)",
"_____no_output_____"
],
[
"#2D Array representing face\nprint(detections)",
"[[305 78 223 223]]\n"
],
[
"for x,y,w,h in detections:\n cv2.rectangle(img,(x,y),(x+w,y+h),(0,255,0),2)\n \ncv2.imshow(\"output\",img)\ncv2.waitKey(0)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b554fd662b25e55b9be5e431872bcebbd16632 | 16,617 | ipynb | Jupyter Notebook | hour-of-callysto-2.ipynb | callysto/hour-of-callysto | cdd206f760cb1f2aaff3e2268ea617f7eaf324ba | [
"MIT"
] | null | null | null | hour-of-callysto-2.ipynb | callysto/hour-of-callysto | cdd206f760cb1f2aaff3e2268ea617f7eaf324ba | [
"MIT"
] | null | null | null | hour-of-callysto-2.ipynb | callysto/hour-of-callysto | cdd206f760cb1f2aaff3e2268ea617f7eaf324ba | [
"MIT"
] | 1 | 2020-01-09T09:24:18.000Z | 2020-01-09T09:24:18.000Z | 29.050699 | 235 | 0.552145 | [
[
[
"%%html\n<style>div.run_this_cell{display:block;}</style>\n<style>table {float:left;width:100%;}</style>",
"_____no_output_____"
]
],
[
[
"<img style=\"float:right;margin-left:50px;margin-right:50px;\" width=\"300\" src=\"images/discovercoding.png\">\n\n# 1. Welcome to the Hour of Callysto!\nLesson created and taught by [Discover Coding](https://discovercoding.ca).\n\nWith support and funding from [Callysto](https://callysto.ca/) and the [Pacific Institute for Mathematical Sciences](https://www.pims.math.ca/)\n",
"_____no_output_____"
],
[
"<img style=\"float:right;padding-left:50px;padding-right:50px;\" width=\"400\" src=\"images/library.jpg\">\n\n## 2. What is Callysto?\n\n<div style=\"padding-top:20px;padding-left:50px;font-size:large;\">\n\n- **Callysto** is a free and online collection of special textbooks for Canadian students. \n\n- It's your own personal school library of the future.\n</div>",
"_____no_output_____"
],
[
"<img style=\"float:left;padding-left:50px;padding-right:10px;margin-right:50px;\" width=\"400px\" src=\"images/notebook.jpg\">\n\n<div style=\"padding-left:50px;padding-top:20px;font-size:large;\">\n\n- Callysto uses **Jupyter** notebooks to display text, images, videos, and even code! \n \n- It's a notebook where you **WRITE** and **RUN** code!\n\n- It's the same tool used in universities by programmers and data scientists!\n</div>",
"_____no_output_____"
],
[
"<img style=\"float:right;padding-right:100px;margin-right:100px;\" width=\"200px\" src=\"images/python.png\">\n\n<div style=\"padding-left:50px;font-size:large;\">\n\n- We are going to use this notebook to learn *coding* with **Python**. \n\n- Python *code* are instructions in a *language* that a computer can understand.\n\n- When we are *coding*, we are telling the computer what to do!\n\n</div>",
"_____no_output_____"
],
[
"\n## 3. Getting Started with Callysto\n\nYou've already made it here! To use Callysto:\n1. Log into Callysto: https://hub.callysto.ca\n1. Find an interesting notebook [from here](https://callysto.ca/learning_modules/)\n1. Click on it to get your own copy of it\n1. Open, read, run it from the hub (look for the .ipynb file)\n",
"_____no_output_____"
],
[
"<img src=\"images/hub.png\">\n",
"_____no_output_____"
],
[
"# 4. Python\n\nWhat makes these notebooks cool is that we can write and run code, such as *Python*, directly inside the notebook! We're not just *READING* a textbook anymore; we can now use it to solve problems for us! \n\n### *Example 4.1*\n\nLet's run our first program using [Turtle graphics](https://en.wikipedia.org/wiki/Turtle_graphics)! Select the code block below and click <img src=\"images/run-button.png\" style=\"display:inline-block;\"> or press `CTRL+ENTER`\n",
"_____no_output_____"
]
],
[
[
"# My First Turtle Program!\nfrom mobilechelonian import Turtle\nted = Turtle()\nted.forward(50)",
"_____no_output_____"
]
],
[
[
"Let's break down what we each line of code that we see:\n\n1. `# My First Turtle Program!` - Any line starting with a `#` is a comment (or notes). It is NOT CODE!\n\n1. `from mobilechelonian import Turtle` - This lets use some `Turtle` code that someone else wrote. We won't worry about it for this class.\n\n1. `ted = Turtle()` - This creates our turtle, named `ted`.\n\n1. `ted.forward(50)` - This tells our turtle `ted` to move `forward` by 50 spaces.",
"_____no_output_____"
],
[
"### *ACTIVITY 4.2*\n\nLet's experiment with the turtle. Change the code above, and re-run it.\nCan you:\n\n1. Change how far `forward` turtle moves. How far can it go?\n1. Can you make turtle move a *negative* number?\n1. Instead of moving `forward`, can you tell turtle to move `backward`?\n1. CHALLENGE 1: Can you rename the turtle from `ted` to a better name?\n1. CHALLENGE 2: Can you tell your turtle to touch BOTH edges of the screen, by only going `backward`?",
"_____no_output_____"
],
[
"## 5. More Turtle *Functions*\n\n`forward()` and `backward()` are called *Functions*. They are commands that a `Turtle()`, like `ted`, understands.\n\nThere are more *functions* that we can use to make turtles do more interesting things.\n\n| Function | What it does |Example| \n|:---|:---|:---|\n| `Turtle.speed(number)` | Set the speed of our turtle, between 1-10 | `t.speed(7)` |\n| `Turtle.right(degrees)` | Turn the turtle `number` of degrees to the right | `t.right(90)` |\n| `Turtle.left(degrees)` | Turn the turtle `number` of degrees to the left | `t.left(90)` |\n| `Turtle.pencolor('color')` | Sets the color of the turtle’s line. <br> The color can be a [color name from this list](https://www.w3schools.com/tags/ref_colornames.asp) | `t.pencolor('Blue')` |\n||||\n \n\n**OBSERVE!!** The functions are applied to `Turtle` objects using the names you gave them.\n\nThe examples above use a Turtle named `t`. You can create more than one turtle, and give them different names!\n\n**Now we can draw more shapes with turtle, and do it faster!**",
"_____no_output_____"
],
[
"### *Example 5.1*\n\nLet's make a 2D shape, like a triangle!\n\nRun the following code:",
"_____no_output_____"
]
],
[
[
"# My Turtle is now 2D!\nfrom mobilechelonian import Turtle\nted = Turtle()\nted.speed(5)\nted.forward(100)\nted.left(120)\nted.forward(100)\nted.left(120)\nted.forward(100)",
"_____no_output_____"
]
],
[
[
"### *ACTIVITY 5.2*\n\nCan you change the color of the triangle?\n\nTry inserting this line of code into Example 5.1:\n\n`ted.pencolor('red')`\n\nTry other [colors named here](https://www.w3schools.com/tags/ref_colornames.asp)\n\n**Observe**\n1. What happens if you write this BEFORE the first move `forward` (on line 5)?\n1. What happens if you write this AFTER the last move `forward` (on line 9)?",
"_____no_output_____"
],
[
"<img style=\"float:right;margin-right:100px;\" width=\"300px\" src=\"images/turtle-house.png\">\n\n### *ACTIVITY 5.3*\n\nCan you make a program where the turtle draws a house that looks like this one?\n\n- **Hint 1:** Start by copy-and-paste from example 5.1\n\n- **Hint 2:** After drawing the triangle, turtle should turn 30 degrees\n\n- **Hint 3:** Draw a line (100 is a good length), turn, draw a line, turn, draw a line...",
"_____no_output_____"
]
],
[
[
"# My Turtle is now 2D!\nfrom mobilechelonian import Turtle\nted = Turtle()\nted.speed(1)\n\n# hint 1 - copy and paste the previous code\n\n# hint 2 - the next turn should be 30 degrees\n\n# hint 3 - forward, turn, forward, turn, forward...",
"_____no_output_____"
]
],
[
[
"# 6. LOOPS\n\n<img style=\"float:right;\" width=\"30%\" src=\"images/loop.gif\">\n\nDid you notice that drawing shapes used the same lines of code over and over?\n\nInstead of typing the same things over and over, we can use a *loop* to run the same code multiple times.\n\nLoops run code __*OVER and OVER and OVER and . . .*__\n\nLoops have two parts:\n1. A line starting with the special keyword `for` or `while`\n1. *indented* lines of code which are run each time\n\n### *Example 6.1*\n\nBelow is an example of a loop using the special keyword `for`. This code says:\n\n- `for` each number called `index`, in the `range` from 0 up to (but not including) 10, move forward, turn right\n\n*Can you predict what it will draw?*\n\nRUN the following code and see!",
"_____no_output_____"
]
],
[
[
"from mobilechelonian import Turtle\nted = Turtle()\nted.speed(5)\n\n# This loop runs (how many?) times\nfor index in range(10): \n ted.forward(50) \n ted.right(80) \n",
"_____no_output_____"
]
],
[
[
"### *ACTIVITY 6.2*\n\nExperiment with the code in Example 6.1, and try different values.\n\nCan you:\n\n1. Make turtle draw faster?\n1. Change how BIG the shape is? (Hint: change the distance turtle moves `forward`)\n1. Change how much the turtle turns each time?\n 1. What happens when turtle turns less than 90?\n 1. What happens when turtle turns 90?\n 1. What happens when turtle turns more than 90?\n1. Can you add some color?",
"_____no_output_____"
],
[
"# 7. RANDOM\n\n<img style=\"float:left;margin-right:30px;\" width=\"400px\" src=\"https://media.giphy.com/media/H4uFElBB9Nt7zq3RZ9/source.gif\">\n\nSo far, we've only been drawing with one color. \n\nLet's make it a little more interesting with using RANDOM colors! \n\nWe'll do 3 things:\n\n1. First, use `import random` in our code.\n1. Next, use `random.randint(0,255)` to pick random numbers from 0 to 255\n1. Last, use the 3 numbers to set a new color using `RGB( number, number, number )`\n\nDon't worry about too much about how this code works.",
"_____no_output_____"
],
[
"### *Example 7.1*\n\nLet's try it out! \n\nRun the following code:",
"_____no_output_____"
]
],
[
[
"from mobilechelonian import Turtle\nimport random\n\nted = Turtle()\nted.speed(10)\n\n# This loop runs 10 times\nfor index in range(10): \n red = random.randint(0,255)\n green = random.randint(0,255)\n blue = random.randint(0,255)\n random_color = \"RGB(%d,%d,%d)\" % (red,green,blue)\n\n ted.pencolor(random_color)\n ted.forward(50) \n ted.right(80) ",
"_____no_output_____"
]
],
[
[
"\n### *ACTIVITY 7.2*\n\nLet's experiment with the code in Example 7.1\n\n1. On line 9, we pick a random number between 0 to 255 for `red`. What happens if we pick a random number between 200 to 255?\n1. What happens if we pick a random number between 0 and 10 for the color `red`?\n1. Try to change the range of random numbers for `green` and `blue`",
"_____no_output_____"
],
[
"\n### *Example 7.3*\n\nLet's try a loop where we draw a SQUARE each time, but do an extra *small* turn before we draw the next square...\n\n",
"_____no_output_____"
]
],
[
[
"from mobilechelonian import Turtle\nimport random\n\nted = Turtle()\nted.speed(10)\n\n# Now we'll run a loop.\nfor index in range(5): \n red = random.randint(0,255)\n green = random.randint(0,255)\n blue = random.randint(0,255)\n random_color = \"RGB(%d,%d,%d)\" % (red,green,blue)\n ted.pencolor(random_color)\n \n ted.forward(100)\n ted.right(90)\n ted.forward(100)\n ted.right(90)\n ted.forward(100)\n ted.right(90)\n ted.forward(100)\n ted.right(90)\n ted.right(20) # Additional small turn before we draw the next square to make a pattern",
"_____no_output_____"
]
],
[
[
"### *ACTIVITY 7.4*\n\nThe picture in Example 7.3 looks incomplete....\n\n1. Can you increase the number of times the loop will run so it'll look better?\n1. Can you make a different pattern just by changing the amount you `right()` turn on line 23?\n1. Can you create a SECOND pattern using another LOOP after the first one completes? \n - HINT 1: COPY and PASTE the ENTIRE LOOP\n - HINT 2: Make sure the second `for` is NOT indented (but the rest of the code IS indented)\n - HINT 3: Change the distance you move `forward()`, and the last `right()` turn",
"_____no_output_____"
],
[
"# 8. SUMMARY\n\nCONGRATULATIONS for making it to the end of this notebook!\n\nToday, you learned:\n\n1. What is Callysto, Jupyter Notebooks, and Python\n1. Wrote a program to draw line art using `turtle`\n1. Used *LOOPS* and *RANDOM* to draw colorful patterns\n\n***\n\n### GREAT JOB! \n### Continue modifying the examples or re-do the activities to make more cool art!\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0b5582d972f5e4e314575d75e465366fd790170 | 731,652 | ipynb | Jupyter Notebook | OptimalDesign/DAS_Designs.ipynb | jbmuir/DAS-Reconstruction | 1cb441010abc350f2fed6418a152e4dc354e92bb | [
"MIT"
] | 1 | 2022-03-02T22:40:38.000Z | 2022-03-02T22:40:38.000Z | OptimalDesign/DAS_Designs.ipynb | jbmuir/DAS-Reconstruction | 1cb441010abc350f2fed6418a152e4dc354e92bb | [
"MIT"
] | null | null | null | OptimalDesign/DAS_Designs.ipynb | jbmuir/DAS-Reconstruction | 1cb441010abc350f2fed6418a152e4dc354e92bb | [
"MIT"
] | null | null | null | 463.364155 | 82,040 | 0.931 | [
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy.io import loadmat\nfrom scipy.interpolate import RectBivariateSpline as rbs\nfrom scipy.integrate import romb\nimport scipy.sparse as sp\nimport os\nimport pywt\nwvt = 'db12'\n%matplotlib inline\nimport matplotlib as mpl\nnorm = mpl.colors.Normalize(vmin=0.0,vmax=1.5)",
"_____no_output_____"
],
[
"nx = ny = 32\nt = np.linspace(0,320,nx+1)\ns = np.linspace(0,320,17)\n\n\nx = y = (t[:-1]+t[1:]) / 2\n\nx = y = (t[:-1]+t[1:]) / 2\nxst = yst = (s[:-1]+s[1:]) / 2\n\nxs, ys = np.meshgrid(xst,yst)\nxs = xs.flatten()\nys = ys.flatten()",
"_____no_output_____"
],
[
"from scipy.signal import butter, lfilter\n\n\ndef butter_bandpass(lowcut, highcut, fs, order=5):\n nyq = 0.5 * fs\n low = lowcut / nyq\n high = highcut / nyq\n b, a = butter(order, [low, high], btype='band')\n return b, a\n\n\ndef butter_bandpass_filter(data, lowcut, highcut, fs, order=5, axis=0):\n b, a = butter_bandpass(lowcut, highcut, fs, order=order)\n y = lfilter(b, a, data, axis=axis)\n return y\n\ndef butter_lowpass(lowcut, fs, order=5):\n nyq = 0.5 * fs\n low = lowcut / nyq\n b, a = butter(order, [low], btype='low')\n return b, a\n\n\ndef butter_low_filter(data, lowcut, fs, order=5, axis=0):\n b, a = butter_lowpass(lowcut, fs, order=order)\n y = lfilter(b, a, data, axis=axis)\n return y",
"_____no_output_____"
],
[
"shot = np.reshape(np.fromfile(\"Testing/TestData/shot1.dat\", dtype=np.float32), (4001,64,64))\nt = np.linspace(0, 0.5, 4001)\nshotf = butter_low_filter(shot, 10, 8000)\ntf = t[::20]\nshotf = shotf[::20,:,:]\ntf_freq = 1/(tf[1]-tf[0])\n\nxc = np.linspace(0,320,65)\nxc = (xc[:-1]+xc[1:])/2\nyc = xc",
"_____no_output_____"
],
[
"shotf_itps = [rbs(xc, yc, s) for s in shotf[:-1]]",
"_____no_output_____"
],
[
"def reconstruction(w, wvt_lens, wvt):\n starts = np.hstack([0,np.cumsum(wvt_lens)])\n wcoef = [w[starts[i]:starts[i+1]] for i in range(len(wvt_lens))]\n return pywt.waverec(wcoef, wvt)",
"_____no_output_____"
]
],
[
[
"# ZigZag",
"_____no_output_____"
]
],
[
[
"das_template_x = np.array([2.5*np.sqrt(2)*i for i in range(24)])\ndas_template_y = np.array([2.5*np.sqrt(2)*i for i in range(24)])\ndas_template_x2 = np.hstack([das_template_x,das_template_x[::-1],das_template_x,das_template_x[::-1]])\ndas_template_y2 = np.hstack([das_template_y,das_template_y+das_template_y[-1],das_template_y+2*das_template_y[-1],das_template_y+3*das_template_y[-1]])\n\ndas_x = np.hstack([das_template_x2+i*das_template_x[-1] for i in range(4)])\ndas_y = np.hstack([das_template_y2 for i in range(4)])\n\noffset = (320-np.max(das_x))/2\n\ndas_x += offset\ndas_y += offset\n\nazimuth_template_1 = np.array([[[45 for i in range(24)], [-45 for i in range(24)]] for i in range(2)]).flatten()\nazimuth_template_2 = np.array([[[135 for i in range(24)], [215 for i in range(24)]] for i in range(2)]).flatten()\ndas_az = np.hstack([azimuth_template_1, azimuth_template_2, \n azimuth_template_1, azimuth_template_2])\n\nraz = np.deg2rad(das_az)\n\n\ncscale = 2\n\ngenerate_kernels = True\n\n\nL = 10 #gauge length\nll = np.linspace(-L/2, L/2, 2**5+1)\ndl = ll[1]-ll[0]\np1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]\np2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]\n\n\nif generate_kernels:\n os.makedirs(\"Kernels\", exist_ok=True)\n crv = loadmat(f\"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat\")\n G_mat = np.reshape(crv[\"G_mat\"].T, (crv[\"G_mat\"].shape[1], nx, ny))\n crvscales = crv[\"scales\"].flatten()\n cvtscaler = 2.0**(cscale*crvscales)\n G1 = np.zeros((len(raz), G_mat.shape[0]))\n G2 = np.zeros((len(raz), G_mat.shape[0]))\n G3 = np.zeros((len(xs), G_mat.shape[0])) \n for j in range(G_mat.shape[0]):\n frame = rbs(x,y,G_mat[j])\n #average derivatives of frame along gauge length\n fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L\n fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L\n G1[:,j] = (np.sin(raz)**2*fd1 + \n np.sin(2*raz)*fd2/2) / cvtscaler[j]\n G2[:,j] = (np.cos(raz)**2*fd2 + \n np.sin(2*raz)*fd1/2) / cvtscaler[j]\n G3[:,j] = frame.ev(xs, ys) / cvtscaler[j]\n\n \n G = np.hstack([G1, G2])\n Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))\n G = G / Gn\n# Gn=1\n G_zigzag = G",
"_____no_output_____"
],
[
"np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))",
"_____no_output_____"
],
[
"plt.plot(np.sort(np.diag(G @ np.linalg.solve(G.T@G + 1e-10*np.eye(G.shape[1]), G.T))))",
"_____no_output_____"
],
[
"exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])\neyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])\nexyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])\nedasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr) \ndas_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])\n\ncuxr = np.array([s.ev(xs, ys, dx=1) for s in shotf_itps])\ncuyr = np.array([s.ev(xs, ys, dy=1) for s in shotf_itps])\n\n\n\nnp.save(\"Testing/zigzag.npy\", das_wvt_data)",
"_____no_output_____"
],
[
"wvt_tmp = pywt.wavedec(edasr.T[0], wvt)\nwvt_lens = [len(wc) for wc in wvt_tmp]",
"_____no_output_____"
],
[
"resi = np.load(f\"Testing/zigzag_res.npy\")\n\nGs = np.std(G)\n\nresxi = resi[:G3.shape[1], :]\nresyi = resi[G3.shape[1]:, :]\n\nxpredi = (G3/Gn/Gs) @ resxi\nypredi = (G3/Gn/Gs) @ resyi\n\n\ntxpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))\ntypredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))\n\n\nres = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))\n",
"_____no_output_____"
],
[
"plt.plot(np.std(resi, axis=1))",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")\n\nplt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))\n, norm=norm)\nplt.colorbar()",
"_____no_output_____"
],
[
"res",
"_____no_output_____"
],
[
"plt.plot(cuxr.T[100])\nplt.plot(txpredi[100])",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,c=das_az)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.colorbar(cax, label=\"Cable Azimuth\")\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")\n",
"_____no_output_____"
],
[
"np.sqrt(np.square(das_x[1:]-das_x[:-1])+np.square(das_y[1:]-das_y[:-1]))",
"_____no_output_____"
]
],
[
[
"# Spiral",
"_____no_output_____"
]
],
[
[
"das_theta2 = np.linspace(0,(360*4)**2, 192*2)\ndas_theta = np.deg2rad(np.sqrt(das_theta2))\na = 0\nb = 1\ndas_r = b*das_theta\n\ndas_x = das_r * np.cos(das_theta)\ndas_y = das_r * np.sin(das_theta)\nraz = np.pi/2-np.arctan2(b*np.tan(das_theta)+(a+b*das_theta), b-(a+b*das_theta)*np.tan(das_theta))\ndas_az = np.rad2deg(raz)\n\nxwidth = np.max(das_x)-np.min(das_x)\ndas_x = das_x / xwidth * 320\ndas_y = das_y / xwidth * 320\nx_offset = 320 - np.max(das_x)\ndas_x = das_x + x_offset\ny_offset = np.min(das_y)\ndas_y = das_y - y_offset\n\nL = 10 #gauge length\nll = np.linspace(-L/2, L/2, 2**5+1)\ndl = ll[1]-ll[0]\np1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]\np2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]\n\n\nif generate_kernels:\n os.makedirs(\"Kernels\", exist_ok=True)\n crv = loadmat(f\"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat\")\n G_mat = np.reshape(crv[\"G_mat\"].T, (crv[\"G_mat\"].shape[1], nx, ny))\n crvscales = crv[\"scales\"].flatten()\n cvtscaler = 2.0**(cscale*crvscales)\n G1 = np.zeros((len(raz), G_mat.shape[0]))\n G2 = np.zeros((len(raz), G_mat.shape[0]))\n for j in range(G_mat.shape[0]):\n frame = rbs(x,y,G_mat[j])\n #average derivatives of frame along gauge length\n fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L\n fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L\n G1[:,j] = (np.sin(raz)**2*fd1 + \n np.sin(2*raz)*fd2/2) / cvtscaler[j]\n G2[:,j] = (np.cos(raz)**2*fd2 + \n np.sin(2*raz)*fd1/2) / cvtscaler[j]\n \n G = np.hstack([G1, G2])\n Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))\n G = G / Gn\n G_spiral = G",
"_____no_output_____"
],
[
"np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))",
"_____no_output_____"
],
[
"exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])\neyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])\nexyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])\nedasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr) \ndas_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])\n\nnp.save(\"Testing/spiral.npy\", das_wvt_data)",
"_____no_output_____"
],
[
"resi = np.load(f\"Testing/spiral_res.npy\")\n\nGs = np.std(G)\n\nresxi = resi[:G3.shape[1], :]\nresyi = resi[G3.shape[1]:, :]\n\nxpredi = (G3/Gn/Gs) @ resxi\nypredi = (G3/Gn/Gs) @ resyi\n\n\ntxpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))\ntypredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))\n\n\nres = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))\n",
"_____no_output_____"
],
[
"res",
"_____no_output_____"
],
[
"plt.plot(cuxr.T[100])\nplt.plot(txpredi[100])",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")\n\nplt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))\n, norm=norm)\nplt.colorbar()",
"_____no_output_____"
],
[
"np.hstack([cuxr, cuyr]).shape",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,c=das_az)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.colorbar(cax, label=\"Cable Azimuth\")\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")\n",
"_____no_output_____"
],
[
"np.sqrt(np.square(das_x[1:]-das_x[:-1])+np.square(das_y[1:]-das_y[:-1]))",
"_____no_output_____"
]
],
[
[
"# Crossing",
"_____no_output_____"
]
],
[
[
"template = np.linspace(0,320, 65)\ntemplate = (template[1:]+template[:-1])/2\n\ndas_x = np.hstack([template, template, template,[80 for i in range(len(template))], [160 for i in range(len(template))], [240 for i in range(len(template))]])\ndas_y = np.hstack([[80 for i in range(len(template))], [160 for i in range(len(template))], [240 for i in range(len(template))],template,template,template])\ndas_az = np.hstack([[90 for i in range(len(template))], [270 for i in range(len(template))], [90 for i in range(len(template))],[0 for i in range(len(template))], [180 for i in range(len(template))], [0 for i in range(len(template))]])\nraz = np.deg2rad(das_az)\n\nL = 10 #gauge length\nll = np.linspace(-L/2, L/2, 2**5+1)\ndl = ll[1]-ll[0]\np1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]\np2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]\n\n\nif generate_kernels:\n os.makedirs(\"Kernels\", exist_ok=True)\n crv = loadmat(f\"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat\")\n G_mat = np.reshape(crv[\"G_mat\"].T, (crv[\"G_mat\"].shape[1], nx, ny))\n crvscales = crv[\"scales\"].flatten()\n cvtscaler = 2.0**(cscale*crvscales)\n G1 = np.zeros((len(raz), G_mat.shape[0]))\n G2 = np.zeros((len(raz), G_mat.shape[0]))\n for j in range(G_mat.shape[0]):\n frame = rbs(x,y,G_mat[j])\n #average derivatives of frame along gauge length\n fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L\n fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L\n G1[:,j] = (np.sin(raz)**2*fd1 + \n np.sin(2*raz)*fd2/2) / cvtscaler[j]\n G2[:,j] = (np.cos(raz)**2*fd2 + \n np.sin(2*raz)*fd1/2) / cvtscaler[j]\n \n G = np.hstack([G1, G2])\n Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))\n G = G / Gn\n G_cross = G",
"_____no_output_____"
],
[
"np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))",
"_____no_output_____"
],
[
"exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])\neyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])\nexyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])\nedasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr) \ndas_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])\n\nnp.save(\"Testing/crossing.npy\", das_wvt_data )",
"_____no_output_____"
],
[
"resi = np.load(f\"Testing/crossing_res.npy\")\n\nGs = np.std(G)\n\nresxi = resi[:G3.shape[1], :]\nresyi = resi[G3.shape[1]:, :]\n\nxpredi = (G3/Gn/Gs) @ resxi\nypredi = (G3/Gn/Gs) @ resyi\n\n\ntxpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))\ntypredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))\n\n\nres = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))\n",
"_____no_output_____"
],
[
"res",
"_____no_output_____"
],
[
"plt.plot(cuxr.T[150])\nplt.plot(txpredi[150])",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")\n\nplt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))\n, norm=norm)\nplt.scatter(xs[150], ys[150], color='r')\nplt.colorbar()",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,c=das_az)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.colorbar(cax, label=\"Cable Azimuth\")\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")",
"_____no_output_____"
]
],
[
[
"# Random",
"_____no_output_____"
]
],
[
[
"template = np.linspace(0,320, 65)\ntemplate = (template[1:]+template[:-1])/2\n\nnp.random.seed(94899109)\ndas_x = np.random.uniform(5,315,384)\ndas_y = np.random.uniform(5,315,384)\ndas_az = np.random.uniform(0,360,384)\n\nraz = np.deg2rad(das_az)\n\nL = 10 #gauge length\nll = np.linspace(-L/2, L/2, 2**5+1)\ndl = ll[1]-ll[0]\np1 = das_x[:,np.newaxis]+np.sin(raz[:,np.newaxis])*ll[np.newaxis,:]\np2 = das_y[:,np.newaxis]+np.cos(raz[:,np.newaxis])*ll[np.newaxis,:]\n\n\nif generate_kernels:\n os.makedirs(\"Kernels\", exist_ok=True)\n crv = loadmat(f\"../Curvelet_Basis_Construction/G_{nx}_{ny}.mat\")\n G_mat = np.reshape(crv[\"G_mat\"].T, (crv[\"G_mat\"].shape[1], nx, ny))\n crvscales = crv[\"scales\"].flatten()\n cvtscaler = 2.0**(cscale*crvscales)\n G1 = np.zeros((len(raz), G_mat.shape[0]))\n G2 = np.zeros((len(raz), G_mat.shape[0]))\n for j in range(G_mat.shape[0]):\n frame = rbs(x,y,G_mat[j])\n #average derivatives of frame along gauge length\n fd1 = romb(frame.ev(p1, p2, dx=1), dl) / L\n fd2 = romb(frame.ev(p1, p2, dy=1), dl) / L\n G1[:,j] = (np.sin(raz)**2*fd1 + \n np.sin(2*raz)*fd2/2) / cvtscaler[j]\n G2[:,j] = (np.cos(raz)**2*fd2 + \n np.sin(2*raz)*fd1/2) / cvtscaler[j]\n \n G = np.hstack([G1, G2])\n Gn = np.max(np.sqrt(np.sum(G**2, axis=1)))\n G = G / Gn\n G_random = G",
"_____no_output_____"
],
[
"np.linalg.slogdet(G.T@G+1e-10*np.eye(G.shape[1]))",
"_____no_output_____"
],
[
"exxr = np.array([romb(s.ev(p1, p2, dx=2), dl)/L for s in shotf_itps])\neyyr = np.array([romb(s.ev(p1, p2, dy=2), dl)/L for s in shotf_itps])\nexyr = np.array([romb(s.ev(p1, p2, dx=1, dy=1), dl)/L for s in shotf_itps])\nedasr = (np.sin(raz)**2*exxr+np.sin(2*raz)*exyr+np.cos(raz)**2*eyyr) \ndas_wvt_data = np.array([np.hstack(pywt.wavedec(d, wvt)) for d in edasr.T])\n\nnp.save(\"Testing/random.npy\", das_wvt_data)",
"_____no_output_____"
],
[
"resi = np.load(f\"Testing/random_res.npy\")\n\nGs = np.std(G)\n\nresxi = resi[:G3.shape[1], :]\nresyi = resi[G3.shape[1]:, :]\n\nxpredi = (G3/Gn/Gs) @ resxi\nypredi = (G3/Gn/Gs) @ resyi\n\n\ntxpredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in xpredi]))\ntypredi = np.real(np.array([reconstruction(w, wvt_lens, wvt) for w in ypredi]))\n\n\nres = np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T), np.square(typredi-cuyr.T)])))/np.std(np.hstack([cuxr, cuyr]))\n",
"_____no_output_____"
],
[
"plt.plot(np.std(resi, axis=1))",
"_____no_output_____"
],
[
"res",
"_____no_output_____"
],
[
"plt.plot(cuxr.T[100])\nplt.plot(txpredi[100])",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,color='k', alpha=0.25)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")\n\nplt.scatter(xs, ys, c= np.sqrt(np.mean(np.hstack([np.square(txpredi-cuxr.T)/np.std(cuxr, axis=0)[:,np.newaxis]**2, np.square(typredi-cuyr.T)/np.std(cuyr, axis=0)[:,np.newaxis]**2]), axis=1))\n, norm=norm)\nplt.colorbar()",
"_____no_output_____"
],
[
"cax = plt.scatter(das_x, das_y,c=das_az)\nplt.xlim(0,320)\nplt.ylim(0,320)\nplt.colorbar(cax, label=\"Cable Azimuth\")\nplt.xlabel(\"Easting (m)\")\nplt.ylabel(\"Northing (m)\")\nplt.gca().set_aspect(\"equal\")",
"_____no_output_____"
],
[
"np.save(\"Kernels/G_zigzag.npy\", G_zigzag)\nnp.save(\"Kernels/G_spiral.npy\", G_spiral)\nnp.save(\"Kernels/G_cross.npy\", G_cross)\nnp.save(\"Kernels/G_random.npy\", G_random)",
"_____no_output_____"
]
],
[
[
"# Eigenvalue Spectrum",
"_____no_output_____"
]
],
[
[
"G_full = np.vstack([np.hstack([G3, np.zeros(G3.shape)]), np.hstack([np.zeros(G3.shape), G3])])\n\nidet = 1e-10*np.eye(G_zigzag.shape[1])\nezig = np.sort(np.linalg.eigvals(G_zigzag.T@G_zigzag+idet))[::-1]\nespi = np.sort(np.linalg.eigvals(G_spiral.T@G_spiral+idet))[::-1]\necro = np.sort(np.linalg.eigvals(G_cross.T@G_cross+idet))[::-1]\neran = np.sort(np.linalg.eigvals(G_random.T@G_random+idet))[::-1]\n# efull= np.sort(np.linalg.eigvals(G_full.T@G_full+idet))[::-1]\n\nezign = ezig / ezig[0]\nespin = espi / espi[0]\necron = ecro / ecro[0]\nerann = eran / eran[0]\n# efulln = efull / efull[0]\n",
"_____no_output_____"
],
[
"plt.plot(np.sort(np.diag(G_zigzag @ np.linalg.solve(G_zigzag.T@G_zigzag + 1e-0*np.eye(G_zigzag.shape[1]), G_zigzag.T))), label=\"ZigZag\")\nplt.plot(np.sort(np.diag(G_spiral @ np.linalg.solve(G_spiral.T@G_spiral + 1e-0*np.eye(G_spiral.shape[1]), G_spiral.T))), label=\"Spiral\")\nplt.plot(np.sort(np.diag(G_cross @ np.linalg.solve(G_cross.T@G_cross + 1e-0*np.eye(G_cross.shape[1]), G_cross.T))), label=\"Crossing\")\nplt.plot(np.sort(np.diag(G_random @ np.linalg.solve(G_random.T@G_random + 1e-0*np.eye(G_random.shape[1]), G_random.T))), label=\"Random\")\n\nplt.xlim(0,384)\nplt.ylabel(\"Coherence\")\nplt.xlabel(\"Sorted Diagonal\")\nplt.legend(loc=\"lower right\")",
"_____no_output_____"
],
[
"plt.plot(np.log10(np.real(ezig)), label=\"ZigZag\")\nplt.plot(np.log10(np.real(espi)), label=\"Spiral\")\nplt.plot(np.log10(np.real(ecro)), label=\"Crossing\")\nplt.plot(np.log10(np.real(eran)), label=\"Random\")\n\nplt.xlim(0,384)\nplt.ylabel(\"Log10 Normalized Eigenvalues\")\nplt.xlabel(\"Eigenvalue Index\")\nplt.legend(loc=\"upper right\")",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0b559bff5e3f5dc802ce48c16073b1512e7cb27 | 27,365 | ipynb | Jupyter Notebook | homework/julia_029_hw.ipynb | h164654156465/1st-JuliaMarathon | f247b3bd50c15b0ca31c134e8c52d824b346ee8b | [
"Apache-2.0"
] | null | null | null | homework/julia_029_hw.ipynb | h164654156465/1st-JuliaMarathon | f247b3bd50c15b0ca31c134e8c52d824b346ee8b | [
"Apache-2.0"
] | null | null | null | homework/julia_029_hw.ipynb | h164654156465/1st-JuliaMarathon | f247b3bd50c15b0ca31c134e8c52d824b346ee8b | [
"Apache-2.0"
] | null | null | null | 52.024715 | 1,989 | 0.370546 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b55a931debb218cd97f6241c10dc97cdc74fa7 | 3,665 | ipynb | Jupyter Notebook | examples/charts/notebook/lines.ipynb | evidation-health/bokeh | 2c580d93419033b962d36e3c46d7606cc2f24606 | [
"BSD-3-Clause"
] | 1 | 2017-08-02T23:12:03.000Z | 2017-08-02T23:12:03.000Z | examples/charts/notebook/lines.ipynb | evidation-health/bokeh | 2c580d93419033b962d36e3c46d7606cc2f24606 | [
"BSD-3-Clause"
] | null | null | null | examples/charts/notebook/lines.ipynb | evidation-health/bokeh | 2c580d93419033b962d36e3c46d7606cc2f24606 | [
"BSD-3-Clause"
] | null | null | null | 28.858268 | 133 | 0.48895 | [
[
[
"import pandas as pd\nfrom bokeh.charts import Line, show, output_notebook, vplot, hplot\nfrom bokeh.charts import defaults",
"_____no_output_____"
],
[
"output_notebook()",
"_____no_output_____"
],
[
"# build a dataset where multiple columns measure the same thing\ndata = dict(python=[2, 3, 7, 5, 26, 221, 44, 233, 254, 265, 266, 267, 120, 111],\n pypy=[12, 33, 47, 15, 126, 121, 144, 233, 254, 225, 226, 267, 110, 130],\n jython=[22, 43, 10, 25, 26, 101, 114, 203, 194, 215, 201, 227, 139, 160],\n test=['foo', 'bar', 'foo', 'bar', 'foo', 'bar', 'foo', 'bar', 'foo', 'bar', 'foo', 'bar', 'foo', 'bar']\n )\ndf = pd.DataFrame(data)\n\n# add a column with a range of dates, as if the values were sampled then\ndf['date'] = pd.date_range('1/1/2015', periods=len(df.index), freq='D')",
"_____no_output_____"
],
[
"# build the line plots\nline0 = Line(df, y=['python', 'pypy', 'jython'],\n title=\"Interpreters (y=['python', 'pypy', 'jython'])\", ylabel='Duration', legend=True)\n\nline1 = Line(df, x='date', y=['python', 'pypy', 'jython'],\n title=\"Interpreters (x='date', y=['python', 'pypy', 'jython'])\", ylabel='Duration', legend=True)\n\nline2 = Line(df, x='date', y=['python', 'pypy', 'jython'],\n dash=['python', 'pypy', 'jython'],\n title=\"Interpreters (x='date', y, dash=['python', 'pypy', 'jython'])\", ylabel='Duration', legend=True)\n\nline3 = Line(df, x='date', y=['python', 'pypy', 'jython'],\n dash=['python', 'pypy', 'jython'],\n color=['python', 'pypy', 'jython'],\n title=\"Interpreters (x='date', y, dash, color=['python', 'pypy', 'jython'])\", ylabel='Duration', legend=True)\n\nline4 = Line(df, x='date', y=['python', 'pypy', 'jython'],\n dash='test',\n color=['python', 'pypy', 'jython'],\n title=\"Interpreters (x='date', y, color=['python', 'pypy', 'jython'], dash='test')\", ylabel='Duration',\n legend=True)",
"_____no_output_____"
],
[
"show(\n vplot(\n hplot(line0, line1),\n hplot(line2, line3),\n hplot(line4)\n )\n)\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0b578f2b0542f752867fe3f10ec76ea92572eb3 | 711,700 | ipynb | Jupyter Notebook | reading_assignments/6_Note-Neural Networks.ipynb | biqar/Fall-2020-ITCS-8156-MachineLearning | ce14609327e5fa13f7af7b904a69da3aa3606f37 | [
"MIT"
] | null | null | null | reading_assignments/6_Note-Neural Networks.ipynb | biqar/Fall-2020-ITCS-8156-MachineLearning | ce14609327e5fa13f7af7b904a69da3aa3606f37 | [
"MIT"
] | null | null | null | reading_assignments/6_Note-Neural Networks.ipynb | biqar/Fall-2020-ITCS-8156-MachineLearning | ce14609327e5fa13f7af7b904a69da3aa3606f37 | [
"MIT"
] | null | null | null | 574.414851 | 118,200 | 0.938934 | [
[
[
"$\\newcommand{\\xv}{\\mathbf{x}}\n \\newcommand{\\wv}{\\mathbf{w}}\n \\newcommand{\\yv}{\\mathbf{y}}\n \\newcommand{\\zv}{\\mathbf{z}}\n \\newcommand{\\Chi}{\\mathcal{X}}\n \\newcommand{\\R}{\\rm I\\!R}\n \\newcommand{\\sign}{\\text{sign}}\n \\newcommand{\\Tm}{\\mathbf{T}}\n \\newcommand{\\Xm}{\\mathbf{X}}\n \\newcommand{\\Xlm}{\\mathbf{X1}}\n \\newcommand{\\Wm}{\\mathbf{W}}\n \\newcommand{\\Vm}{\\mathbf{V}}\n \\newcommand{\\Ym}{\\mathbf{Y}}\n \\newcommand{\\Zm}{\\mathbf{Z}}\n \\newcommand{\\Zlm}{\\mathbf{Z1}}\n \\newcommand{\\I}{\\mathbf{I}}\n \\newcommand{\\muv}{\\boldsymbol\\mu}\n \\newcommand{\\Sigmav}{\\boldsymbol\\Sigma}\n \\newcommand{\\Phiv}{\\boldsymbol\\Phi}\n$\n\n# Neural Networks\n\nNeural networks, or artificial neural networks, are the computational models inspired by the brain. Mimicing the neurons' synaptic connecions (Figure 1), we build or stack multiple neuron-like hidden units to map data into nonlinear space for rich representation. \n\n<img src=\"https://upload.wikimedia.org/wikipedia/commons/1/10/Blausen_0657_MultipolarNeuron.png\" width=500/>\n<center>Figure 1. Anatomy of a neuron (wikipedia) </center>\n\nNow, let us review the perceptron model. \n\n<img src=\"http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/perceptron.png\" width=600 />\n\nIn perceptron, passing the output of linear model to the step function, we get discrete outputs. \nNow, you can think a perceptron as a neuron. With a threshold zero, when the linear model outputs are over it, it passes the signal to next neuron. \n\nBy connecting the perceptrons, we can actually build synaptic connections.\nWe call this model as *multi-layer perceptron* (MLP). \n\n",
"_____no_output_____"
],
[
"**Q:** For inputs $x \\in \\{-1, +1 \\}$, think about what the following picture represents and answer for it. \n\n1)\n<img src=\"http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_q1.png\" width=300/>\n \n2)\n<img src=\"http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_q2.png\" width=300/>\n\n3)\n<img src=\"http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_q3.png\" width=700/>\n",
"_____no_output_____"
],
[
"Answer: \n\n1) +1\n\n2) -1\n\n3) -1",
"_____no_output_____"
],
[
"## Feed Forward Neural Networks\n\nFitting the data with MLP is a combinatorial optimization problem with non-smooth step function. \nSo, we can consider smooth step function, a s-shaped sigmoid function. \nWe call this smooth function as **activation function**.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline \n\nfig, ax = plt.subplots()\n\n# x - y axis\nax.axhline(y=0, color='k', linewidth=1)\nax.axvline(x=0, color='k', linewidth=1)\n\n# step function in blue \nplt.plot([0, 6], [1, 1], 'b-', linewidth=3)\nplt.plot([-6, 0], [-1, -1], 'b-', linewidth=3)\nplt.plot([0, 0], [-1, 1], 'b-', linewidth=3)\n\n# tanh in red\nx = np.linspace(-6, 6, 100)\nplt.plot(x, np.tanh(x), 'r-', linewidth=3)\n",
"_____no_output_____"
]
],
[
[
"## Non-linear Extension of Linear Model\n\nAs we discussed, feed forward neural networks have a rich representation. Thus, it can represent the linear model with single layer. \n\n<img src=\"http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/mlp_linear.png\" width=400/>\n\nConsidering the multiple outputs, we formulated this in matrix: \n\n$$\n\\begin{align}\nE &= \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{k=1}^{K} (t_{nk} - y_{nk})^2 \\\\\n\\\\\n\\Ym &= \\Xlm \\cdot \\Wm\n\\end{align}\n$$\n\nHere, we assume the first column of $\\Xlm$ is the bias column with 1's. \nThus, the weight matrix $\\Wm$ is $(D+1) \\times K$ with the bias row in the first row. \n\nFrom this model, we can convert the raw data $\\Xm$ to $\\Phiv$, which is a nonlinear mapping.\n\n$$\n\\phi: \\Xm \\rightarrow \\Phiv\n$$\n\nThen, we can rewrite the linear model with as follows:\n\n$$\n\\begin{align}\nE &= \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{k=1}^{K} (t_{nk} - y_{nk})^2 \n\\\\\n\\Ym &= \\Phiv \\Wm \\\\ \n\\\\\n\\Ym_{nk} &= \\Phiv_n^\\top \\Wm_k \n\\end{align}\n$$\n\nNow, let $\\phi(\\xv) = h(\\xv)$ where $h$ is the *activation function*. \n\n$$\n\\begin{align}\n\\Zm &= h(\\Xlm \\cdot \\Vm) \\\\\n\\\\\n\\Ym & = \\Zlm \\cdot \\Wm \n\\end{align}\n$$\n\nFigure below depics this model. \n\n<img src=\"http://webpages.uncc.edu/mlee173/teach/itcs4156online/images/class/nn.png\" width=500/>\n\nThe size of each matrix is listed: \n- $\\Xm: N \\times D$\n- $\\Xlm: N \\times (D+1)$\n- $\\Vm: (D+1) \\times G$\n- $\\Zm: N \\times G$\n- $\\Zlm: N \\times (G+1)$\n- $\\Wm: (G+1) \\times K$\n- $\\Ym: N \\times K$\n\nFor this two-layer network, we call the blue circle layer with the activation functions as **hidden layer** and the organge layer with summation as **output layer**.",
"_____no_output_____"
],
[
"# Why Sigmoid? \n\nThe resemblance to the step function can be good reason. But is there any other reason for choosing a sigmoid function as activation? \n\nLet us take a look at a polinomial function and the sigmoid.\n\n$$\ny = x^4 + 3 x^2 + 7 x + 3 \\quad\\quad\\text{vs.}\\quad\\quad y = tanh(x)\n$$",
"_____no_output_____"
]
],
[
[
"# polinomial function\ndef h_poly(x): \n return x**4 + 3 * x**2 + 7 * x + 3\n\n# sigmoid function\ndef h_sigmoid(x): \n return np.tanh(x)\n\n##### Gradient functions\n\n# polinomial function\ndef dh_poly(x): \n return 4 * x**3 + 6 * x + 7\n\n# polinomial function\ndef dh_sigmoid(x): \n h = h_sigmoid(x)\n return 1 - h ** 2\n\nx = np.linspace(-6, 6, 100)\n\nplt.figure(figsize=(16,8))\nplt.subplot(121)\nplt.plot(x, h_poly(x), label=\"$y = x^4 + 3 x^2 + 7 x + 3$\")\nplt.plot(x, dh_poly(x), label=\"$dy$\")\nplt.legend()\n\nplt.subplot(122)\nplt.plot(x, h_sigmoid(x), label=\"$y = tanh(x)$\")\nplt.plot(x, dh_sigmoid(x), label=\"$dy$\")\nplt.legend()\n",
"_____no_output_____"
]
],
[
[
"Here, we can see the polinomial gradients are very huge when $x$ is moving away from 0. A gradient descent procedure takes this huge step for the large positive or negative $x$ values, which can make learning divergent and unstable.\n\nIn the right figure, we can see the gradient is nearly turned off for large $x$ values. Only on the nonlinear region of sigmoid function, small gradient is applied for stable learning. ",
"_____no_output_____"
],
[
"# Gradient Descent\n\nFrom the error function $E$, \n\n$$\nE = \\frac{1}{N} \\frac{1}{K}\\sum_{n=1}^{N} \\sum_{k=1}^{K} (t_{nk} - y_{nk})^2,\n$$\n\nwe can derive the gradient to update the weights for each layer. \n\nSince we can change the output and eventually the error by changing the weights $\\Vm$ and $\\Wm$, \n\n$$\n\\begin{align}\nv_{dg} &\\leftarrow v_{dg} - \\alpha_h \\frac{\\partial{E}} {\\partial{v_{dg}}} \\\\ \n\\\\ \nw_{gk} &\\leftarrow w_{gk} - \\alpha_o \\frac{\\partial{E}} {\\partial{w_{gk}}},\n\\end{align}\n$$\n\nwhere $\\alpha_h$ and $\\alpha_o$ are the learning rate for hidden and output layer respectively. \n\n$$\n\\begin{align}\n\\frac{\\partial{E}}{\\partial{w_{gk}}} &= \\frac{\\partial{\\Big( \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl})^2} \\Big)}{\\partial{w_{gk}}} \\\\\n &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N}(t_{nk} - y_{nk}) \\frac{\\partial{y_{nl}}}{\\partial{w_{gk}}} \n\\end{align}\n$$\n\nwhere \n\n$$\ny_{nl} = z1_{n}^\\top w_{*l} = \\sum_{g=0}^{G} z1_{ng} w_{gl} . \n$$\n\nThe gradient for the output layer can be computed as follows:\n$$\n\\begin{align}\n\\frac{\\partial{E}}{\\partial{w_{gk}}} &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} (t_{nk} - y_{nk}) z1_{nk} \\\\\n &= -2 \\frac{1}{N} \\frac{1}{K} \\Zlm^\\top (\\Tm - \\Ym).\n\\end{align} \n$$\n\nFor the hidden layer, \n\n$$\n\\begin{align}\n\\frac{\\partial{E}}{\\partial{v_{dg}}} &= \\frac{\\partial{\\Big( \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl})^2} \\Big)}{\\partial{v_{dg}}} \\\\\n &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl}) \\frac{\\partial{y_{nl}}}{\\partial{v_{dg}}} \n\\end{align}\n$$\n\nwhere \n\n$$\ny_{nl} = \\sum_{g=0}^{G} z1_{ng} w_{gl} = \\sum_{g=0}^G w_{gl} h (\\sum_{d=0}^D v_{dg} x1_{nd}) . \n$$\n\nLet $a_{ng} = \\sum_{d=0}^D x1_{nd} v_{dg}$. Then, we can use a chain rule for the derivation. \n\n$$\n\\begin{align}\n\\frac{\\partial{E}}{\\partial{v_{dg}}} &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl}) \\frac{\\partial{\\Big( \\sum_{q=0}^G w_{ql} h (\\sum_{p=0}^D v_{pq} x1_{np}) \\Big)}}{\\partial{v_{dg}}} \\\\\n &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl}) \\sum_{q=0}^G w_{ql} \\frac{\\partial{\\Big( h (\\sum_{p=0}^D v_{pq} x1_{np}) \\Big)}}{\\partial{v_{dg}}} \\\\\n &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl}) \\sum_{q=0}^G w_{ql} \\frac{\\partial{h(a_{ng})}}{\\partial{a_{ng}}} \\frac{\\partial{a_{ng}}}{\\partial{v_{dg}}} \\\\\n &= -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nl} - y_{nl}) \\sum_{q=0}^G w_{ql} \\frac{\\partial{h(a_{ng})}}{\\partial{a_{ng}}} x1_{nd}.\n\\end{align}\n$$\n\nWhen $h = tanh$, \n\n$$\n\\frac{\\partial{h(a_{ng})}}{\\partial{a_{ng}}} = \\frac{z_{ng}}{\\partial{a_{ng}}} = (1 - z_{ng}^2). \n$$\n\nThus, \n\n$$\n\\frac{\\partial{E}}{\\partial{v_{dg}}} = -2 \\frac{1}{N} \\frac{1}{K} \\sum_{n=1}^{N} \\sum_{l=1}^{K} (t_{nk} - y_{nl}) \\sum_{g=0}^G w_{gl} (1 - z_{ng}^2) x1_{nd}.\n$$\n\nRewriting this in matrix form, \n\n$$\n\\frac{\\partial{E}}{\\partial{v_{dg}}} = -2 \\frac{1}{N} \\frac{1}{K} \\Xlm^\\top \\Big( (\\Tm - \\Ym) \\Wm^\\top \\odot (1 - \\Zm^2) \\Big).\n$$\n\nHere, $\\odot$ denotes the element-wise multiplication.\n\nTo summarize, the backpropagation performs the this weight updates iteratively: \n$$\n\\begin{align}\n\\Vm &\\leftarrow \\Vm + \\rho_h \\frac{1}{N} \\frac{1}{K} \\Xlm^\\top \\Big( (\\Tm - \\Ym) \\Wm^\\top \\odot (1 - \\Zm^2) \\Big), \\\\\n\\Wm &\\leftarrow \\Wm + \\rho_o \\frac{1}{N} \\frac{1}{K} \\Zlm^\\top \\Big( \\Tm - \\Ym \\Big)\n\\end{align}\n$$\nwhere $\\rho_h$ and $\\rho_o$ are the learning rate for hidden and output layer weights. \n\nImplemented iteration follows. ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nimport IPython.display as ipd # for display and clear_output\nimport time # for sleep",
"_____no_output_____"
],
[
"# Make some training data\nn = 20\nX = np.linspace(0.,20.0,n).reshape((n,1)) - 10\nT = 0.2 + 0.05 * (X+10) + 0.4 * np.sin(X+10) + 0.2 * np.random.normal(size=(n,1))\n\n# Make some testing data\nXtest = X + 0.1*np.random.normal(size=(n,1))\nTtest = 0.2 + 0.05 * (Xtest+10) + 0.4 * np.sin(Xtest+10) + 0.2 * np.random.normal(size=(n,1))\n\nnSamples = X.shape[0]\nnOutputs = T.shape[1]",
"_____no_output_____"
],
[
"# Set parameters of neural network\nnHiddens = 10\n\nrhoh = 0.5\nrhoo = 0.1\n\nrh = rhoh / (nSamples*nOutputs)\nro = rhoo / (nSamples*nOutputs)\n\n# Initialize weights to uniformly distributed values between small normally-distributed between -0.1 and 0.1\nV = 0.1*2*(np.random.uniform(size=(1+1,nHiddens))-0.5)\nW = 0.1*2*(np.random.uniform(size=(1+nHiddens,nOutputs))-0.5)\n\n# Add constant column of 1's\ndef addOnes(A):\n return np.insert(A, 0, 1, axis=1)\nX1 = addOnes(X)\nXtest1 = addOnes(Xtest)\n\n# Take nReps steepest descent steps in gradient descent search in mean-squared-error function\nnReps = 30000\n# collect training and testing errors for plotting\nerrorTrace = np.zeros((nReps,2))\n\nN_ = X1.shape[0]\nK_ = W.shape[1]\n\nfig = plt.figure(figsize=(10,8))\nfor reps in range(nReps):\n\n # Forward pass on training data\n Z = np.tanh(X1 @ V)\n Z1 = addOnes(Z)\n Y = Z1 @ W\n\n # Error in output\n error = T - Y\n \n print(\"V:\", V.shape)\n print(\"X1:\", X1.T.shape)\n print(\"error:\", error.shape)\n print(\"W.T:\", W.T.shape)\n print(\"Z:\", Z.shape)\n print(np.square(Z).shape)\n\n # TODO: Backward pass - the backpropagation and weight update steps\n V = V + ((rh / (N_ * K_)) * X1.T * ((error * W.T) @ (1 - np.square(Z))))\n W = W + (ro / (N_ * K_)) * Z1.T * error\n\n # error traces for plotting\n errorTrace[reps,0] = np.sqrt(np.mean((error**2)))\n Ytest = addOnes(np.tanh(Xtest1 @ V)) @ W #!! Forward pass in one line\n errorTrace[reps,1] = np.sqrt(np.mean((Ytest-Ttest)**2))\n\n if reps % 1000 == 0 or reps == nReps-1:\n plt.clf()\n plt.subplot(3,1,1)\n plt.plot(errorTrace[:reps,:])\n plt.ylim(0,0.7)\n plt.xlabel('Epochs')\n plt.ylabel('RMSE')\n plt.legend(('Train','Test'),loc='upper left')\n \n plt.subplot(3,1,2)\n plt.plot(X,T,'o-',Xtest,Ttest,'o-',Xtest,Ytest,'o-')\n plt.xlim(-10,10)\n plt.legend(('Training','Testing','Model'),loc='upper left')\n plt.xlabel('$x$')\n plt.ylabel('Actual and Predicted $f(x)$')\n \n plt.subplot(3,1,3)\n plt.plot(X,Z)\n plt.ylim(-1.1,1.1)\n plt.xlabel('$x$')\n plt.ylabel('Hidden Unit Outputs ($z$)');\n \n ipd.clear_output(wait=True)\n ipd.display(fig)\nipd.clear_output(wait=True)",
"V: (2, 10)\nX1: (2, 20)\nerror: (20, 1)\nW.T: (1, 11)\nZ: (20, 10)\n(20, 10)\n"
]
],
[
[
"$\\newcommand{\\xv}{\\mathbf{x}}\n \\newcommand{\\wv}{\\mathbf{w}}\n \\newcommand{\\yv}{\\mathbf{y}}\n \\newcommand{\\zv}{\\mathbf{z}}\n \\newcommand{\\av}{\\mathbf{a}}\n \\newcommand{\\Chi}{\\mathcal{X}}\n \\newcommand{\\R}{\\rm I\\!R}\n \\newcommand{\\sign}{\\text{sign}}\n \\newcommand{\\Tm}{\\mathbf{T}}\n \\newcommand{\\Xm}{\\mathbf{X}}\n \\newcommand{\\Xlm}{\\mathbf{X1}}\n \\newcommand{\\Wm}{\\mathbf{W}}\n \\newcommand{\\Vm}{\\mathbf{V}}\n \\newcommand{\\Ym}{\\mathbf{Y}}\n \\newcommand{\\Zm}{\\mathbf{Z}}\n \\newcommand{\\Zlm}{\\mathbf{Z1}}\n \\newcommand{\\I}{\\mathbf{I}}\n \\newcommand{\\muv}{\\boldsymbol\\mu}\n \\newcommand{\\Sigmav}{\\boldsymbol\\Sigma}\n \\newcommand{\\Phiv}{\\boldsymbol\\Phi}\n$\n\n# Optimization\n\nSo far, we have been using gradient descent to find minimum or maximum values in our error function. \nIn general, we call this maximization or minimization problem as an **optimization problem**. \n\nIn optimization problems, we look for the largest or the smallest value that a function can take. By systematically choosing input vales within the constraint set, optimization problem seeks for the best available values of an objective function.\n\nSo, for a given function $f(x)$ that maps $f: \\Xm \\rightarrow \\Ym $ where $\\Ym \\subset \\R$, \nwe are looking for a $x^* \\in \\Xm$ that satisfies \n\n$$\n\\begin{cases}\n f(x^*) \\le f(x) &\\forall x & \\quad \\text{if } \\text{ minimization}\\\\\n f(x^*) \\ge f(x) &\\forall x & \\quad \\text{if } \\text{ maximization}.\n \\end{cases}\n$$\n \nThe optimization problems are often expressed in following notation.\n\n$$\n\\begin{equation*}\n\\begin{aligned}\n& \\underset{x}{\\text{minimize}}\n& & f(x) \\\\\n& \\text{subject to}\n& & x \\le b_i, \\; i = 1, \\ldots, m,\\\\\n &&& x \\ge 0.\n\\end{aligned}\n\\end{equation*}\n$$",
"_____no_output_____"
],
[
"## Least Squares\n\n$$\n\\begin{equation*}\n\\begin{aligned}\n& \\underset{\\wv}{\\text{minimize}}\n& & \\Vert \\Xm \\wv - t\\Vert^2\n\\end{aligned}\n\\end{equation*}\n$$\n\n\n\n\nAs we discussed, least-squares problems can be solved analytically, $\\wv = (\\Xm^\\top \\Xm)^{-1} \\Xm^\\top t$.\nWe easily formulate least-sqaures and solve very efficiently. \n\n## Linear Programming\n\n$$\n\\begin{equation*}\n\\begin{aligned}\n& \\underset{\\xv}{\\text{minimize}}\n& & \\wv^\\top \\xv \\\\\n& \\text{subject to}\n& & \\av_i^\\top \\xv \\le b_i, \\; i = 1, \\ldots, m.\n\\end{aligned}\n\\end{equation*}\n$$\n\n\n\nLinear programming or linear optimization finds a maximum or minimum from a mathematical model that is represented by linear relationships. \nThere is no analytical formular for a solution, but there are reliable algorithms that solve LP efficiently. \n\n\n## Convex Optimization\n\n$$\n\\begin{equation*}\n\\begin{aligned}\n& \\underset{x}{\\text{minimize}}\n& & f_0(x) \\\\\n& \\text{subject to}\n& & f_i(x) \\leq b_i, \\; i = 1, \\ldots, m.\n\\end{aligned}\n\\end{equation*}\n$$\n\n\n\nConvex condition: \n$$\nf_i(\\alpha x_1 + (1-\\alpha) x_2) \\le \\alpha f_i(x_1) + (1-\\alpha) f_i(x_2)\n$$\n\n\nConvex optimization generalizes the linear programming problems. A convex optimization problem has the constraint set that forms convex functions. As a general model of LP, convex optimization problems do not have analytical solution but they also have reliable and efficient algorithms for it. Thus, it can be solved very quickly and reliably up to very large problems.\nHowever, it is difficulty to recognize if it is convex or not.\n\n\n## Nonlinear Optimization\n\nFor non-convex problems, we can apply local optimization methods, which is called nonlinear programming. \nStarting from initial guess, it searchs for a minimal point near neighborhood. It can be fast and can be applicable large problems. However, there is no guarantee for discovery of global optimum. ",
"_____no_output_____"
],
[
"## Newton's method\n\nNewton's method approximates the curve with quadratic function repeatedly to find a temporary point or stationary point of $f$. \n\nIf we assume that for each measurement point $x^{(k)}$, we can compute $f(x^{(k)})$, $f^{\\prime}(x^{(k)})$, and $f^{\\prime\\prime}(x^{(k)})$.\nUsing second order Taylor expansion, we can approximate $q(x)$ for $f(x + \\Delta x)$:\n\n$$\nq(x) = f(x^{(k)}) + f^{\\prime}(x^{(k)}) \\Delta x + \\frac{1}{2} f^{\\prime\\prime}(x^{(k)}) \\Delta x^2\n$$\n\nwhere $\\Delta x = (x - x^{(k)})$. \n\nMinimizing this quadratic function, \n\n$$\n0 = q^\\prime(x) = f^{\\prime}(x^{(k)}) + f^{\\prime\\prime}(x^{(k)}) \\Delta x. \n$$\n\nSetting $x = x^{(k+1)}$, we can get\n\n$$\n x^{(k+1)} = x^{(k)} - \\frac{f^{\\prime}(x^{(k)})}{f^{\\prime\\prime}(x^{(k)})}.\n$$",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport scipy.optimize as opt\nfrom scipy.optimize import rosen, minimize\n",
"_____no_output_____"
],
[
"# examples are from http://people.duke.edu/~ccc14/sta-663-2017/14C_Optimization_In_Python.html\n\nx = np.linspace(-5, 5, 1000)\ny = np.linspace(-5, 5, 1000)\n\nxs, ys = np.meshgrid(x, y)\nzs = rosen(np.vstack([xs.ravel(), ys.ravel()])).reshape(xs.shape)\n\nplt.figure(figsize=(8,8))\nplt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')\nplt.text(1, 1, 'x', va='center', ha='center', color='k', fontsize=30);",
"_____no_output_____"
],
[
"from scipy.optimize import rosen_der, rosen_hess\n\ndef reporter(p):\n \"\"\"record the points visited\"\"\"\n global ps\n ps.append(p)\n\n# starting position\nx0 = np.array([4,-4.1])\n\nps = [x0]\nminimize(rosen, x0, method=\"Newton-CG\", jac=rosen_der, hess=rosen_hess, callback=reporter)\n",
"_____no_output_____"
],
[
"ps = np.array(ps)\nplt.figure(figsize=(16, 8))\nplt.subplot(121)\nplt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')\nplt.plot(ps[:, 0], ps[:, 1], '-ro')\nplt.subplot(122)\nplt.semilogy(range(len(ps)), rosen(ps.T));",
"_____no_output_____"
]
],
[
[
"## Vs. others?\n\nNow, let us take a look at other optimization tools including naive steepest descent and scaled conjugate gradient ([Moller, 1997](http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.50.8063&rep=rep1&type=pdf)). \nTo run this properly, you need to download [grad.py](https://webpages.uncc.edu/mlee173/teach/itcs4156online/notes/grad.py) under your current work folder. ",
"_____no_output_____"
]
],
[
[
"from grad import steepest\n\nres = steepest(np.array(x0), rosen_der, rosen, stepsize=0.0001, wtracep=True, ftracep=True)\n\nps = np.array(res['wtrace'])\nplt.figure(figsize=(16, 8))\nplt.subplot(121)\nplt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')\nplt.plot(ps[:, 0], ps[:, 1], '-ro')\nplt.subplot(122)\nplt.semilogy(range(len(ps)), res['ftrace']);",
"_____no_output_____"
],
[
"from grad import scg\n\nres = scg(np.array(x0), rosen_der, rosen, wtracep=True, ftracep=True)\nres1 = scg(np.array([-4, 4]), rosen_der, rosen, wtracep=True, ftracep=True)\n\n\nps = np.array(res['wtrace'])\nps1 = np.array(res1['wtrace'])\nplt.figure(figsize=(16, 8))\nplt.subplot(121)\nplt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')\nplt.plot(ps[:, 0], ps[:, 1], '-ro')\nplt.plot(ps1[:, 0], ps1[:, 1], '-bo')\nplt.subplot(122)\nplt.semilogy(range(len(ps)), res['ftrace']);",
"_____no_output_____"
],
[
"x0 = [-4, 4]\nps = [x0]\nminimize(rosen, x0, method=\"Newton-CG\", jac=rosen_der, hess=rosen_hess, callback=reporter)\n\nps = np.array(ps)\nplt.figure(figsize=(16, 8))\nplt.subplot(121)\nplt.contour(xs, ys, zs, np.arange(10)**5, cmap='jet')\nplt.plot(ps[:, 0], ps[:, 1], '-ro')\nplt.subplot(122)\nplt.semilogy(range(len(ps)), rosen(ps.T));",
"_____no_output_____"
]
],
[
[
"# Neural Network! \n\nNow, let us use this optimization trick for our neural networks. \n",
"_____no_output_____"
]
],
[
[
"# standardization class\nclass Standardizer: \n \"\"\" class version of standardization \"\"\"\n def __init__(self, X, explore=False):\n self._mu = np.mean(X,0) \n self._sigma = np.std(X,0)\n if explore:\n print (\"mean: \", self._mu)\n print (\"sigma: \", self._sigma)\n print (\"min: \", np.min(X,0))\n print (\"max: \", np.max(X,0))\n\n def set_sigma(self, s):\n self._sigma[:] = s\n\n def standardize(self,X):\n return (X - self._mu) / self._sigma \n\n def unstandardize(self,X):\n return (X * self._sigma) + self._mu \n ",
"_____no_output_____"
],
[
"\n\"\"\" Neural Network \n referenced NN code by Chuck Anderson in R and C++ \n\n by Jake Lee (lemin)\n\n example usage:\n X = numpy.array([0,0,1,0,0,1,1,1]).reshape(4,2)\n T = numpy.array([0,1,1,0,1,0,0,1]).reshape(4,2)\n\n nn = nnet.NeuralNet([2,3,2])\n nn.train(X,T, wprecision=1e-20, fprecision=1e-2)\n Y = nn.use(X)\n\n\"\"\"\nfrom grad import scg, steepest\nfrom copy import copy\n\n\nclass NeuralNet:\n \"\"\" neural network class for regression\n \n Parameters\n ----------\n nunits: list\n the number of inputs, hidden units, and outputs\n\n Methods\n -------\n set_hunit \n update/initiate weights\n\n pack \n pack multiple weights of each layer into one vector\n\n forward\n forward processing of neural network\n\n backward\n back-propagation of neural network\n\n train\n train the neural network\n\n use\n appply the trained network for prediction\n\n Attributes\n ----------\n _nLayers\n the number of hidden unit layers \n\n rho\n learning rate\n\n _W\n weights\n _weights\n weights in one dimension (_W is referencing _weight)\n\n stdX\n standardization class for data\n stdT\n standardization class for target\n\n Notes\n -----\n \n \"\"\"\n\n \n # TODO: Try to implement Neural Network class with the member variables and methods described above\n \n \n \n \n \n \n ",
"_____no_output_____"
],
[
"X = np.array([0,0,1,0,0,1,1,1]).reshape(4,2)\nT = np.array([0,1,1,0,1,0,0,1]).reshape(4,2)\n\nnn = NeuralNet([2,3,2])\nnn.train(X, T) \nY = nn.use(X)",
"_____no_output_____"
],
[
"Y",
"_____no_output_____"
],
[
"T",
"_____no_output_____"
],
[
"X",
"_____no_output_____"
],
[
"# repeating the previous example\n\n# Make some training data\nn = 20\nX = np.linspace(0.,20.0,n).reshape((n,1)) - 10\nT = 0.2 + 0.05 * (X+10) + 0.4 * np.sin(X+10) + 0.2 * np.random.normal(size=(n,1))\n\n# Make some testing data\nXtest = X + 0.1*np.random.normal(size=(n,1))\nTtest = 0.2 + 0.05 * (Xtest+10) + 0.4 * np.sin(Xtest+10) + 0.2 * np.random.normal(size=(n,1))\n\nnSamples = X.shape[0]\nnOutputs = T.shape[1]",
"_____no_output_____"
],
[
"nn = NeuralNet([1,3,1])\nnn.train(X, T, ftracep=True) \nYtest, Z = nn.use(Xtest, retZ=True)\n\nplt.figure(figsize=(10,8))\nplt.subplot(3,1,1)\nplt.plot(nn.ftrace)\nplt.ylim(0,0.7)\nplt.xlabel('Epochs')\nplt.ylabel('RMSE')\nplt.legend(('Train','Test'),loc='upper left')\n\nplt.subplot(3,1,2)\nplt.plot(X,T,'o-',Xtest,Ttest,'o-',Xtest,Ytest,'o-')\nplt.xlim(-10,10)\nplt.legend(('Training','Testing','Model'),loc='upper left')\nplt.xlabel('$x$')\nplt.ylabel('Actual and Predicted $f(x)$')\n\nplt.subplot(3,1,3)\nplt.plot(X, Z[1])\nplt.ylim(-1.1,1.1)\nplt.xlabel('$x$')\nplt.ylabel('Hidden Unit Outputs ($z$)');",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b5967641a2ef5147d8b4a20d651e60d41ca79f | 34,112 | ipynb | Jupyter Notebook | week07_seq2seq/practice_torch.ipynb | dmgirdyuk/Practical_RL | caa8baa385d29f4999386f4c1d47f8d4efbdec01 | [
"Unlicense"
] | 1 | 2020-08-03T21:04:16.000Z | 2020-08-03T21:04:16.000Z | week07_seq2seq/practice_torch.ipynb | dmgirdyuk/Practical_RL | caa8baa385d29f4999386f4c1d47f8d4efbdec01 | [
"Unlicense"
] | null | null | null | week07_seq2seq/practice_torch.ipynb | dmgirdyuk/Practical_RL | caa8baa385d29f4999386f4c1d47f8d4efbdec01 | [
"Unlicense"
] | null | null | null | 38.807736 | 271 | 0.604685 | [
[
[
"## Reinforcement Learning for seq2seq\n\nThis time we'll solve a problem of transribing hebrew words in english, also known as g2p (grapheme2phoneme)\n\n * word (sequence of letters in source language) -> translation (sequence of letters in target language)\n\nUnlike what most deep learning practicioners do, we won't only train it to maximize likelihood of correct translation, but also employ reinforcement learning to actually teach it to translate with as few errors as possible.\n\n\n### About the task\n\nOne notable property of Hebrew is that it's consonant language. That is, there are no wovels in the written language. One could represent wovels with diacritics above consonants, but you don't expect people to do that in everyay life.\n\nTherefore, some hebrew characters will correspond to several english letters and others - to none, so we should use encoder-decoder architecture to figure that out.\n\n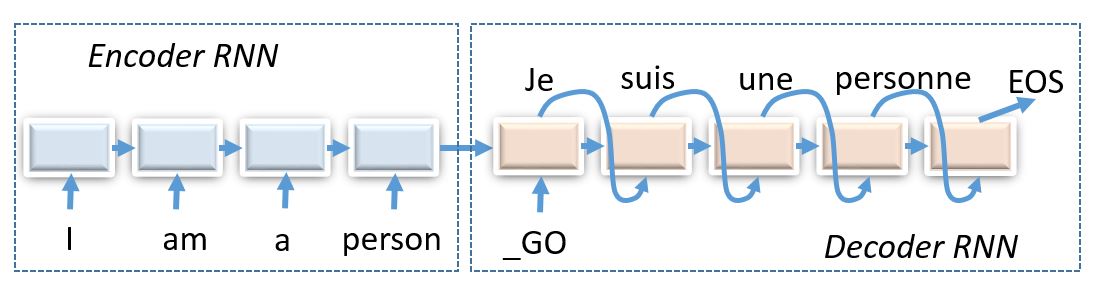\n_(img: esciencegroup.files.wordpress.com)_\n\nEncoder-decoder architectures are about converting anything to anything, including\n * Machine translation and spoken dialogue systems\n * [Image captioning](http://mscoco.org/dataset/#captions-challenge2015) and [image2latex](https://htmlpreview.github.io/?https://github.com/openai/requests-for-research/blob/master/_requests_for_research/im2latex.html) (convolutional encoder, recurrent decoder)\n * Generating [images by captions](https://arxiv.org/abs/1511.02793) (recurrent encoder, convolutional decoder)\n * Grapheme2phoneme - convert words to transcripts\n \nWe chose simplified __Hebrew->English__ machine translation for words and short phrases (character-level), as it is relatively quick to train even without a gpu cluster.",
"_____no_output_____"
]
],
[
[
"import sys\nif 'google.colab' in sys.modules:\n !wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/basic_model_torch.py -O basic_model_torch.py\n !wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/main_dataset.txt -O main_dataset.txt\n !wget https://raw.githubusercontent.com/yandexdataschool/Practical_RL/spring20/week07_seq2seq/voc.py -O voc.py\n !pip3 install torch==1.0.0 nltk editdistance",
"_____no_output_____"
],
[
"# If True, only translates phrases shorter than 20 characters (way easier).\nEASY_MODE = True\n# Useful for initial coding.\n# If false, works with all phrases (please switch to this mode for homework assignment)\n\n# way we translate. Either \"he-to-en\" or \"en-to-he\"\nMODE = \"he-to-en\"\n# maximal length of _generated_ output, does not affect training\nMAX_OUTPUT_LENGTH = 50 if not EASY_MODE else 20\nREPORT_FREQ = 100 # how often to evaluate validation score",
"_____no_output_____"
]
],
[
[
"### Step 1: preprocessing\n\nWe shall store dataset as a dictionary\n`{ word1:[translation1,translation2,...], word2:[...],...}`.\n\nThis is mostly due to the fact that many words have several correct translations.\n\nWe have implemented this thing for you so that you can focus on more interesting parts.\n\n\n__Attention python2 users!__ You may want to cast everything to unicode later during homework phase, just make sure you do it _everywhere_.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom collections import defaultdict\nword_to_translation = defaultdict(list) # our dictionary\n\nbos = '_'\neos = ';'\n\nwith open(\"main_dataset.txt\", encoding=\"utf-8\") as fin:\n for line in fin:\n\n en, he = line[:-1].lower().replace(bos, ' ').replace(eos,\n ' ').split('\\t')\n word, trans = (he, en) if MODE == 'he-to-en' else (en, he)\n\n if len(word) < 3:\n continue\n if EASY_MODE:\n if max(len(word), len(trans)) > 20:\n continue\n\n word_to_translation[word].append(trans)\n\nprint(\"size = \", len(word_to_translation))",
"_____no_output_____"
],
[
"# get all unique lines in source language\nall_words = np.array(list(word_to_translation.keys()))\n# get all unique lines in translation language\nall_translations = np.array(list(set(\n [ts for all_ts in word_to_translation.values() for ts in all_ts])))",
"_____no_output_____"
]
],
[
[
"### split the dataset\n\nWe hold out 10% of all words to be used for validation.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\ntrain_words, test_words = train_test_split(\n all_words, test_size=0.1, random_state=42)",
"_____no_output_____"
]
],
[
[
"### Building vocabularies\n\nWe now need to build vocabularies that map strings to token ids and vice versa. We're gonna need these fellas when we feed training data into model or convert output matrices into english words.",
"_____no_output_____"
]
],
[
[
"from voc import Vocab\ninp_voc = Vocab.from_lines(''.join(all_words), bos=bos, eos=eos, sep='')\nout_voc = Vocab.from_lines(''.join(all_translations), bos=bos, eos=eos, sep='')",
"_____no_output_____"
],
[
"# Here's how you cast lines into ids and backwards.\nbatch_lines = all_words[:5]\nbatch_ids = inp_voc.to_matrix(batch_lines)\nbatch_lines_restored = inp_voc.to_lines(batch_ids)\n\nprint(\"lines\")\nprint(batch_lines)\nprint(\"\\nwords to ids (0 = bos, 1 = eos):\")\nprint(batch_ids)\nprint(\"\\nback to words\")\nprint(batch_lines_restored)",
"_____no_output_____"
]
],
[
[
"Draw word/translation length distributions to estimate the scope of the task.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline\nplt.figure(figsize=[8, 4])\nplt.subplot(1, 2, 1)\nplt.title(\"words\")\nplt.hist(list(map(len, all_words)), bins=20)\n\nplt.subplot(1, 2, 2)\nplt.title('translations')\nplt.hist(list(map(len, all_translations)), bins=20)",
"_____no_output_____"
]
],
[
[
"### Step 3: deploy encoder-decoder (1 point)\n\n__assignment starts here__\n\nOur architecture consists of two main blocks:\n* Encoder reads words character by character and outputs code vector (usually a function of last RNN state)\n* Decoder takes that code vector and produces translations character by character\n\nThan it gets fed into a model that follows this simple interface:\n* __`model(inp, out, **flags) -> logp`__ - takes symbolic int32 matrices of hebrew words and their english translations. Computes the log-probabilities of all possible english characters given english prefices and hebrew word.\n* __`model.translate(inp, **flags) -> out, logp`__ - takes symbolic int32 matrix of hebrew words, produces output tokens sampled from the model and output log-probabilities for all possible tokens at each tick.\n * if given flag __`greedy=True`__, takes most likely next token at each iteration. Otherwise samples with next token probabilities predicted by model.\n\nThat's all! It's as hard as it gets. With those two methods alone you can implement all kinds of prediction and training.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"from basic_model_torch import BasicTranslationModel\nmodel = BasicTranslationModel(inp_voc, out_voc,\n emb_size=64, hid_size=256)",
"_____no_output_____"
],
[
"# Play around with symbolic_translate and symbolic_score\ninp = torch.tensor(np.random.randint(0, 10, [3, 5]), dtype=torch.int64)\nout = torch.tensor(np.random.randint(0, 10, [3, 5]), dtype=torch.int64)\n\n# translate inp (with untrained model)\nsampled_out, logp = model.translate(inp, greedy=False)\n\nprint(\"Sample translations:\\n\", sampled_out)\nprint(\"Log-probabilities at each step:\\n\", logp)",
"_____no_output_____"
],
[
"# score logp(out | inp) with untrained input\nlogp = model(inp, out)\nprint(\"Symbolic_score output:\\n\", logp)\n\nprint(\"Log-probabilities of output tokens:\\n\",\n torch.gather(logp, dim=2, index=out[:, :, None]))",
"_____no_output_____"
],
[
"def translate(lines, max_len=MAX_OUTPUT_LENGTH):\n \"\"\"\n You are given a list of input lines. \n Make your neural network translate them.\n :return: a list of output lines\n \"\"\"\n # Convert lines to a matrix of indices\n lines_ix = inp_voc.to_matrix(lines)\n lines_ix = torch.tensor(lines_ix, dtype=torch.int64)\n\n # Compute translations in form of indices\n trans_ix = <YOUR CODE>\n\n # Convert translations back into strings\n return out_voc.to_lines(trans_ix.data.numpy())",
"_____no_output_____"
],
[
"print(\"Sample inputs:\", all_words[:3])\nprint(\"Dummy translations:\", translate(all_words[:3]))\ntrans = translate(all_words[:3])\n\nassert translate(all_words[:3]) == translate(\n all_words[:3]), \"make sure translation is deterministic (use greedy=True and disable any noise layers)\"\nassert type(translate(all_words[:3])) is list and (type(translate(all_words[:1])[0]) is str or type(\n translate(all_words[:1])[0]) is unicode), \"translate(lines) must return a sequence of strings!\"\n# note: if translation freezes, make sure you used max_len parameter\nprint(\"Tests passed!\")",
"_____no_output_____"
]
],
[
[
"### Scoring function\n\nLogLikelihood is a poor estimator of model performance.\n* If we predict zero probability once, it shouldn't ruin entire model.\n* It is enough to learn just one translation if there are several correct ones.\n* What matters is how many mistakes model's gonna make when it translates!\n\nTherefore, we will use minimal Levenshtein distance. It measures how many characters do we need to add/remove/replace from model translation to make it perfect. Alternatively, one could use character-level BLEU/RougeL or other similar metrics.\n\nThe catch here is that Levenshtein distance is not differentiable: it isn't even continuous. We can't train our neural network to maximize it by gradient descent.",
"_____no_output_____"
]
],
[
[
"import editdistance # !pip install editdistance\n\n\ndef get_distance(word, trans):\n \"\"\"\n A function that takes word and predicted translation\n and evaluates (Levenshtein's) edit distance to closest correct translation\n \"\"\"\n references = word_to_translation[word]\n assert len(references) != 0, \"wrong/unknown word\"\n return min(editdistance.eval(trans, ref) for ref in references)\n\n\ndef score(words, bsize=100):\n \"\"\"a function that computes levenshtein distance for bsize random samples\"\"\"\n assert isinstance(words, np.ndarray)\n\n batch_words = np.random.choice(words, size=bsize, replace=False)\n batch_trans = translate(batch_words)\n\n distances = list(map(get_distance, batch_words, batch_trans))\n\n return np.array(distances, dtype='float32')",
"_____no_output_____"
],
[
"# should be around 5-50 and decrease rapidly after training :)\n[score(test_words, 10).mean() for _ in range(5)]",
"_____no_output_____"
]
],
[
[
"## Step 2: Supervised pre-training (2 points)\n\nHere we define a function that trains our model through maximizing log-likelihood a.k.a. minimizing crossentropy.",
"_____no_output_____"
]
],
[
[
"import random\n\n\ndef sample_batch(words, word_to_translation, batch_size):\n \"\"\"\n sample random batch of words and random correct translation for each word\n example usage:\n batch_x,batch_y = sample_batch(train_words, word_to_translations,10)\n \"\"\"\n # choose words\n batch_words = np.random.choice(words, size=batch_size)\n\n # choose translations\n batch_trans_candidates = list(map(word_to_translation.get, batch_words))\n batch_trans = list(map(random.choice, batch_trans_candidates))\n return batch_words, batch_trans",
"_____no_output_____"
],
[
"bx, by = sample_batch(train_words, word_to_translation, batch_size=3)\nprint(\"Source:\")\nprint(bx)\nprint(\"Target:\")\nprint(by)",
"_____no_output_____"
],
[
"from basic_model_torch import infer_length, infer_mask, to_one_hot\n\n\ndef compute_loss_on_batch(input_sequence, reference_answers):\n \"\"\" Compute crossentropy loss given a batch of sources and translations \"\"\"\n input_sequence = torch.tensor(inp_voc.to_matrix(input_sequence), dtype=torch.int64)\n reference_answers = torch.tensor(out_voc.to_matrix(reference_answers), dtype=torch.int64)\n\n # Compute log-probabilities of all possible tokens at each step. Use model interface.\n logprobs_seq = <YOUR CODE>\n\n # compute elementwise crossentropy as negative log-probabilities of reference_answers.\n crossentropy = - \\\n torch.sum(logprobs_seq *\n to_one_hot(reference_answers, len(out_voc)), dim=-1)\n assert crossentropy.dim(\n ) == 2, \"please return elementwise crossentropy, don't compute mean just yet\"\n\n # average with mask\n mask = infer_mask(reference_answers, out_voc.eos_ix)\n loss = torch.sum(crossentropy * mask) / torch.sum(mask)\n\n return loss",
"_____no_output_____"
],
[
"# test it\nloss = compute_loss_on_batch(*sample_batch(train_words, word_to_translation, 3))\nprint('loss = ', loss)\n\nassert loss.item() > 0.0\nloss.backward()\nfor w in model.parameters():\n assert w.grad is not None and torch.max(torch.abs(w.grad)).item() != 0, \\\n \"Loss is not differentiable w.r.t. a weight with shape %s. Check comput_loss_on_batch.\" % (\n w.size(),)",
"_____no_output_____"
]
],
[
[
"##### Actually train the model\n\nMinibatches and stuff...",
"_____no_output_____"
]
],
[
[
"from IPython.display import clear_output\nfrom tqdm import tqdm, trange # or use tqdm_notebook,tnrange\n\nloss_history = []\neditdist_history = []\nentropy_history = []\nopt = torch.optim.Adam(model.parameters())",
"_____no_output_____"
],
[
"\n\nfor i in trange(25000):\n loss = compute_loss_on_batch(*sample_batch(train_words, word_to_translation, 32))\n\n # train with backprop\n loss.backward()\n opt.step()\n opt.zero_grad()\n\n loss_history.append(loss.item())\n\n if (i+1) % REPORT_FREQ == 0:\n clear_output(True)\n current_scores = score(test_words)\n editdist_history.append(current_scores.mean())\n print(\"llh=%.3f, mean score=%.3f\" %\n (np.mean(loss_history[-10:]), np.mean(editdist_history[-10:])))\n plt.figure(figsize=(12, 4))\n plt.subplot(131)\n plt.title('train loss / traning time')\n plt.plot(loss_history)\n plt.grid()\n plt.subplot(132)\n plt.title('val score distribution')\n plt.hist(current_scores, bins=20)\n plt.subplot(133)\n plt.title('val score / traning time (lower is better)')\n plt.plot(editdist_history)\n plt.grid()\n plt.show()",
"_____no_output_____"
]
],
[
[
"__How to interpret the plots:__\n\n* __Train loss__ - that's your model's crossentropy over minibatches. It should go down steadily. Most importantly, it shouldn't be NaN :)\n* __Val score distribution__ - distribution of translation edit distance (score) within batch. It should move to the left over time.\n* __Val score / training time__ - it's your current mean edit distance. This plot is much whimsier than loss, but make sure it goes below 8 by 2500 steps. \n\nIf it doesn't, first try to re-create both model and opt. You may have changed it's weight too much while debugging. If that doesn't help, it's debugging time.",
"_____no_output_____"
]
],
[
[
"for word in train_words[:10]:\n print(\"%s -> %s\" % (word, translate([word])[0]))",
"_____no_output_____"
],
[
"test_scores = []\nfor start_i in trange(0, len(test_words), 32):\n batch_words = test_words[start_i:start_i+32]\n batch_trans = translate(batch_words)\n distances = list(map(get_distance, batch_words, batch_trans))\n test_scores.extend(distances)\n\nprint(\"Supervised test score:\", np.mean(test_scores))",
"_____no_output_____"
]
],
[
[
"## Self-critical policy gradient (2 points)\n\nIn this section you'll implement algorithm called self-critical sequence training (here's an [article](https://arxiv.org/abs/1612.00563)).\n\nThe algorithm is a vanilla policy gradient with a special baseline. \n\n$$ \\nabla J = E_{x \\sim p(s)} E_{y \\sim \\pi(y|x)} \\nabla log \\pi(y|x) \\cdot (R(x,y) - b(x)) $$\n\nHere reward R(x,y) is a __negative levenshtein distance__ (since we minimize it). The baseline __b(x)__ represents how well model fares on word __x__.\n\nIn practice, this means that we compute baseline as a score of greedy translation, $b(x) = R(x,y_{greedy}(x)) $.\n\n\n\n\nLuckily, we already obtained the required outputs: `model.greedy_translations, model.greedy_mask` and we only need to compute levenshtein using `compute_levenshtein` function.\n",
"_____no_output_____"
]
],
[
[
"def compute_reward(input_sequence, translations):\n \"\"\" computes sample-wise reward given token ids for inputs and translations \"\"\"\n distances = list(map(get_distance,\n inp_voc.to_lines(input_sequence.data.numpy()),\n out_voc.to_lines(translations.data.numpy())))\n # use negative levenshtein distance so that larger reward means better policy\n return - torch.tensor(distances, dtype=torch.int64)",
"_____no_output_____"
],
[
"def scst_objective_on_batch(input_sequence, max_len=MAX_OUTPUT_LENGTH):\n \"\"\" Compute pseudo-loss for policy gradient given a batch of sources \"\"\"\n input_sequence = torch.tensor(inp_voc.to_matrix(input_sequence), dtype=torch.int64)\n\n # use model to __sample__ symbolic translations given input_sequence\n sample_translations, sample_logp = <YOUR CODE>\n # use model to __greedy__ symbolic translations given input_sequence\n greedy_translations, greedy_logp = <YOUR CODE>\n\n # compute rewards and advantage\n rewards = compute_reward(input_sequence, sample_translations)\n baseline = <YOUR CODE: compute __negative__ levenshtein for greedy mode>\n\n # compute advantage using rewards and baseline\n advantage = <YOUR CODE>\n\n # compute log_pi(a_t|s_t), shape = [batch, seq_length]\n logp_sample = <YOUR CODE>\n \n # ^-- hint: look at how crossentropy is implemented in supervised learning loss above\n # mind the sign - this one should not be multiplied by -1 :)\n\n # policy gradient pseudo-loss. Gradient of J is exactly policy gradient.\n J = logp_sample * advantage[:, None]\n\n assert J.dim() == 2, \"please return elementwise objective, don't compute mean just yet\"\n\n # average with mask\n mask = infer_mask(sample_translations, out_voc.eos_ix)\n loss = - torch.sum(J * mask) / torch.sum(mask)\n\n # regularize with negative entropy. Don't forget the sign!\n # note: for entropy you need probabilities for all tokens (sample_logp), not just logp_sample\n entropy = <YOUR CODE: compute entropy matrix of shape[batch, seq_length], H = -sum(p*log_p), don't forget the sign!>\n # hint: you can get sample probabilities from sample_logp using math :)\n\n assert entropy.dim(\n ) == 2, \"please make sure elementwise entropy is of shape [batch,time]\"\n\n reg = - 0.01 * torch.sum(entropy * mask) / torch.sum(mask)\n\n return loss + reg, torch.sum(entropy * mask) / torch.sum(mask)",
"_____no_output_____"
]
],
[
[
"# Policy gradient training\n",
"_____no_output_____"
]
],
[
[
"entropy_history = [np.nan] * len(loss_history)\nopt = torch.optim.Adam(model.parameters(), lr=1e-5)",
"_____no_output_____"
],
[
"for i in trange(100000):\n loss, ent = scst_objective_on_batch(\n sample_batch(train_words, word_to_translation, 32)[0]) # [0] = only source sentence\n\n # train with backprop\n loss.backward()\n opt.step()\n opt.zero_grad()\n\n loss_history.append(loss.item())\n entropy_history.append(ent.item())\n\n if (i+1) % REPORT_FREQ == 0:\n clear_output(True)\n current_scores = score(test_words)\n editdist_history.append(current_scores.mean())\n plt.figure(figsize=(12, 4))\n plt.subplot(131)\n plt.title('val score distribution')\n plt.hist(current_scores, bins=20)\n plt.subplot(132)\n plt.title('val score / traning time')\n plt.plot(editdist_history)\n plt.grid()\n plt.subplot(133)\n plt.title('policy entropy / traning time')\n plt.plot(entropy_history)\n plt.grid()\n plt.show()\n print(\"J=%.3f, mean score=%.3f\" %\n (np.mean(loss_history[-10:]), np.mean(editdist_history[-10:])))",
"_____no_output_____"
]
],
[
[
"__Debugging tips:__\n<img src=https://github.com/yandexdataschool/Practical_RL/raw/master/yet_another_week/_resource/do_something_scst.png width=400>\n\n * As usual, don't expect improvements right away, but in general the model should be able to show some positive changes by 5k steps.\n * Entropy is a good indicator of many problems. \n * If it reaches zero, you may need greater entropy regularizer.\n * If it has rapid changes time to time, you may need gradient clipping.\n * If it oscillates up and down in an erratic manner... it's perfectly okay for entropy to do so. But it should decrease at the end.\n \n * We don't show loss_history cuz it's uninformative for pseudo-losses in policy gradient. However, if something goes wrong you can check it to see if everything isn't a constant zero.",
"_____no_output_____"
],
[
"### Results",
"_____no_output_____"
]
],
[
[
"for word in train_words[:10]:\n print(\"%s -> %s\" % (word, translate([word])[0]))",
"_____no_output_____"
],
[
"test_scores = []\nfor start_i in trange(0, len(test_words), 32):\n batch_words = test_words[start_i:start_i+32]\n batch_trans = translate(batch_words)\n distances = list(map(get_distance, batch_words, batch_trans))\n test_scores.extend(distances)\nprint(\"Supervised test score:\", np.mean(test_scores))\n\n# ^^ If you get Out Of MemoryError, please replace this with batched computation",
"_____no_output_____"
]
],
[
[
"## Step 6: Make it actually work (5++ pts)\n\nIn this section we want you to finally __restart with EASY_MODE=False__ and experiment to find a good model/curriculum for that task.\n\nWe recommend you to start with the following architecture\n\n```\nencoder---decoder\n\n P(y|h)\n ^\n LSTM -> LSTM\n ^ ^\n biLSTM -> LSTM\n ^ ^\ninput y_prev\n```\n\n__Note:__ you can fit all 4 state tensors of both LSTMs into a in a single state - just assume that it contains, for example, [h0, c0, h1, c1] - pack it in encode and update in decode.\n\n\nHere are some cool ideas on what you can do then.\n\n__General tips & tricks:__\n* You will likely need to adjust pre-training time for such a network.\n* Supervised pre-training may benefit from clipping gradients somehow.\n* SCST may indulge a higher learning rate in some cases and changing entropy regularizer over time.\n* It's often useful to save pre-trained model parameters to not re-train it every time you want new policy gradient parameters. \n* When leaving training for nighttime, try setting REPORT_FREQ to a larger value (e.g. 500) not to waste time on it.\n\n__Formal criteria:__\nTo get 5 points we want you to build an architecture that:\n* _doesn't consist of single GRU_\n* _works better_ than single GRU baseline. \n* We also want you to provide either learning curve or trained model, preferably both\n* ... and write a brief report or experiment log describing what you did and how it fared.\n\n### Attention\nThere's more than one way to connect decoder to encoder\n * __Vanilla:__ layer_i of encoder last state goes to layer_i of decoder initial state\n * __Every tick:__ feed encoder last state _on every iteration_ of decoder.\n * __Attention:__ allow decoder to \"peek\" at one (or several) positions of encoded sequence on every tick.\n \nThe most effective (and cool) of those is, of course, attention.\nYou can read more about attention [in this nice blog post](https://distill.pub/2016/augmented-rnns/). The easiest way to begin is to use \"soft\" attention with \"additive\" or \"dot-product\" intermediate layers.\n\n__Tips__\n* Model usually generalizes better if you no longer allow decoder to see final encoder state\n* Once your model made it through several epochs, it is a good idea to visualize attention maps to understand what your model has actually learned\n\n* There's more stuff [here](https://github.com/yandexdataschool/Practical_RL/blob/master/week8_scst/bonus.ipynb)\n* If you opted for hard attention, we recommend [gumbel-softmax](https://blog.evjang.com/2016/11/tutorial-categorical-variational.html) instead of sampling. Also please make sure soft attention works fine before you switch to hard.\n\n### UREX\n* This is a way to improve exploration in policy-based settings. The main idea is that you find and upweight under-appreciated actions.\n* Here's [video](https://www.youtube.com/watch?v=fZNyHoXgV7M&feature=youtu.be&t=3444)\n and an [article](https://arxiv.org/abs/1611.09321).\n* You may want to reduce batch size 'cuz UREX requires you to sample multiple times per source sentence.\n* Once you got it working, try using experience replay with importance sampling instead of (in addition to) basic UREX.\n\n### Some additional ideas:\n* (advanced deep learning) It may be a good idea to first train on small phrases and then adapt to larger ones (a.k.a. training curriculum).\n* (advanced nlp) You may want to switch from raw utf8 to something like unicode or even syllables to make task easier.\n* (advanced nlp) Since hebrew words are written __with vowels omitted__, you may want to use a small Hebrew vowel markup dataset at `he-pron-wiktionary.txt`.\n\n",
"_____no_output_____"
]
],
[
[
"assert not EASY_MODE, \"make sure you set EASY_MODE = False at the top of the notebook.\"",
"_____no_output_____"
]
],
[
[
"`[your report/log here or anywhere you please]`",
"_____no_output_____"
],
[
"__Contributions:__ This notebook is brought to you by\n* Yandex [MT team](https://tech.yandex.com/translate/)\n* Denis Mazur ([DeniskaMazur](https://github.com/DeniskaMazur)), Oleg Vasilev ([Omrigan](https://github.com/Omrigan/)), Dmitry Emelyanenko ([TixFeniks](https://github.com/tixfeniks)) and Fedor Ratnikov ([justheuristic](https://github.com/justheuristic/))\n* Dataset is parsed from [Wiktionary](https://en.wiktionary.org), which is under CC-BY-SA and GFDL licenses.\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0b59791dc8db48b7a01905d011c1c866e188126 | 17,732 | ipynb | Jupyter Notebook | nbs/lr_scheduler.ipynb | IamGianluca/petfinder-pawpularity-score | 955471a031ac14f7447f5ecf1b1a2423073f1bf4 | [
"MIT"
] | null | null | null | nbs/lr_scheduler.ipynb | IamGianluca/petfinder-pawpularity-score | 955471a031ac14f7447f5ecf1b1a2423073f1bf4 | [
"MIT"
] | null | null | null | nbs/lr_scheduler.ipynb | IamGianluca/petfinder-pawpularity-score | 955471a031ac14f7447f5ecf1b1a2423073f1bf4 | [
"MIT"
] | null | null | null | 197.022222 | 15,692 | 0.913772 | [
[
[
"import transformers \nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"total_samples = 9000\nbs = 32\nn_epochs = 10\n\nnum_warmup_steps = (total_samples // bs) * 2\nnum_total_steps = (total_samples // bs) * n_epochs\n\nmodel = nn.Linear(2, 1)\noptimizer = optim.SGD(model.parameters(), lr=0.01)\nscheduler = transformers.get_cosine_schedule_with_warmup(optimizer, \n num_warmup_steps=num_warmup_steps, \n num_training_steps=num_total_steps)\nlrs = []\nfor i in range(num_total_steps):\n optimizer.step()\n lrs.append(optimizer.param_groups[0][\"lr\"])\n scheduler.step()\n \nplt.plot(lrs)\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0b5a6e9a6927dca671a11e7b4e4dd99738e13fe | 23,160 | ipynb | Jupyter Notebook | ipython-notebooks/notebooks/libraries/DEAP.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 1 | 2021-12-13T15:41:48.000Z | 2021-12-13T15:41:48.000Z | ipython-notebooks/notebooks/libraries/DEAP.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 15 | 2021-09-12T15:06:13.000Z | 2022-03-31T19:02:08.000Z | ipython-notebooks/notebooks/libraries/DEAP.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
] | 1 | 2022-01-29T00:37:52.000Z | 2022-01-29T00:37:52.000Z | 33.613933 | 369 | 0.538687 | [
[
[
"# DEAP",
"_____no_output_____"
],
[
"DEAP is a novel evolutionary computation framework for rapid prototyping and testing of ideas. It seeks to make algorithms explicit and data structures transparent. It works in perfect harmony with parallelisation mechanism such as multiprocessing and SCOOP. The following documentation presents the key concepts and many features to build your own evolutions.\n\nLibrary documentation: <a>http://deap.readthedocs.org/en/master/</a>",
"_____no_output_____"
],
[
"## One Max Problem (GA)",
"_____no_output_____"
],
[
"This problem is very simple, we search for a 1 filled list individual. This problem is widely used in the evolutionary computation community since it is very simple and it illustrates well the potential of evolutionary algorithms.",
"_____no_output_____"
]
],
[
[
"import random\n\nfrom deap import base\nfrom deap import creator\nfrom deap import tools",
"_____no_output_____"
],
[
"# creator is a class factory that can build new classes at run-time\ncreator.create(\"FitnessMax\", base.Fitness, weights=(1.0,))\ncreator.create(\"Individual\", list, fitness=creator.FitnessMax)",
"_____no_output_____"
],
[
"# a toolbox stores functions and their arguments\ntoolbox = base.Toolbox()\n\n# attribute generator\ntoolbox.register(\"attr_bool\", random.randint, 0, 1)\n\n# structure initializers\ntoolbox.register(\"individual\", tools.initRepeat, creator.Individual, toolbox.attr_bool, 100)\ntoolbox.register(\"population\", tools.initRepeat, list, toolbox.individual)",
"_____no_output_____"
],
[
"# evaluation function\ndef evalOneMax(individual):\n return sum(individual),",
"_____no_output_____"
],
[
"# register the required genetic operators\ntoolbox.register(\"evaluate\", evalOneMax)\ntoolbox.register(\"mate\", tools.cxTwoPoint)\ntoolbox.register(\"mutate\", tools.mutFlipBit, indpb=0.05)\ntoolbox.register(\"select\", tools.selTournament, tournsize=3)",
"_____no_output_____"
],
[
"random.seed(64)\n\n# instantiate a population\npop = toolbox.population(n=300)\nCXPB, MUTPB, NGEN = 0.5, 0.2, 40\n\n# evaluate the entire population\nfitnesses = list(map(toolbox.evaluate, pop))\nfor ind, fit in zip(pop, fitnesses):\n ind.fitness.values = fit\n\nprint(\" Evaluated %i individuals\" % len(pop))",
" Evaluated 300 individuals\n"
],
[
"# begin the evolution\nfor g in range(NGEN):\n print(\"-- Generation %i --\" % g)\n\n # select the next generation individuals\n offspring = toolbox.select(pop, len(pop))\n\n # clone the selected individuals\n offspring = list(map(toolbox.clone, offspring))\n\n # apply crossover and mutation on the offspring\n for child1, child2 in zip(offspring[::2], offspring[1::2]):\n if random.random() < CXPB:\n toolbox.mate(child1, child2)\n del child1.fitness.values\n del child2.fitness.values\n\n for mutant in offspring:\n if random.random() < MUTPB:\n toolbox.mutate(mutant)\n del mutant.fitness.values\n\n # evaluate the individuals with an invalid fitness\n invalid_ind = [ind for ind in offspring if not ind.fitness.valid]\n fitnesses = map(toolbox.evaluate, invalid_ind)\n for ind, fit in zip(invalid_ind, fitnesses):\n ind.fitness.values = fit\n\n print(\" Evaluated %i individuals\" % len(invalid_ind))\n\n # the population is entirely replaced by the offspring\n pop[:] = offspring\n\n # gather all the fitnesses in one list and print the stats\n fits = [ind.fitness.values[0] for ind in pop]\n\n length = len(pop)\n mean = sum(fits) / length\n sum2 = sum(x*x for x in fits)\n std = abs(sum2 / length - mean**2)**0.5\n\n print(\" Min %s\" % min(fits))\n print(\" Max %s\" % max(fits))\n print(\" Avg %s\" % mean)\n print(\" Std %s\" % std)",
"-- Generation 0 --\n Evaluated 189 individuals\n Min 40.0\n Max 65.0\n Avg 54.7433333333\n Std 4.46289766358\n-- Generation 1 --\n Evaluated 171 individuals\n Min 44.0\n Max 70.0\n Avg 58.48\n Std 3.98533980149\n-- Generation 2 --\n Evaluated 169 individuals\n Min 54.0\n Max 68.0\n Avg 61.6066666667\n Std 2.92779021714\n-- Generation 3 --\n Evaluated 185 individuals\n Min 57.0\n Max 73.0\n Avg 63.82\n Std 2.74364720764\n-- Generation 4 --\n Evaluated 175 individuals\n Min 54.0\n Max 73.0\n Avg 65.67\n Std 2.57961883489\n-- Generation 5 --\n Evaluated 164 individuals\n Min 60.0\n Max 76.0\n Avg 67.5466666667\n Std 2.57833710407\n-- Generation 6 --\n Evaluated 185 individuals\n Min 63.0\n Max 77.0\n Avg 69.0666666667\n Std 2.50510589707\n-- Generation 7 --\n Evaluated 194 individuals\n Min 62.0\n Max 78.0\n Avg 70.78\n Std 2.39963886172\n-- Generation 8 --\n Evaluated 199 individuals\n Min 63.0\n Max 79.0\n Avg 72.3133333333\n Std 2.57717330077\n-- Generation 9 --\n Evaluated 169 individuals\n Min 67.0\n Max 81.0\n Avg 74.0\n Std 2.62551582234\n-- Generation 10 --\n Evaluated 180 individuals\n Min 67.0\n Max 83.0\n Avg 75.9166666667\n Std 2.52910831893\n-- Generation 11 --\n Evaluated 193 individuals\n Min 67.0\n Max 84.0\n Avg 77.5966666667\n Std 2.40291258453\n-- Generation 12 --\n Evaluated 177 individuals\n Min 72.0\n Max 85.0\n Avg 78.97\n Std 2.29690371297\n-- Generation 13 --\n Evaluated 195 individuals\n Min 70.0\n Max 86.0\n Avg 80.13\n Std 2.35650164439\n-- Generation 14 --\n Evaluated 175 individuals\n Min 74.0\n Max 86.0\n Avg 81.3966666667\n Std 2.03780655499\n-- Generation 15 --\n Evaluated 181 individuals\n Min 74.0\n Max 87.0\n Avg 82.33\n Std 2.18504767301\n-- Generation 16 --\n Evaluated 198 individuals\n Min 74.0\n Max 88.0\n Avg 83.4033333333\n Std 2.22575580172\n-- Generation 17 --\n Evaluated 190 individuals\n Min 72.0\n Max 88.0\n Avg 84.14\n Std 2.34955314901\n-- Generation 18 --\n Evaluated 170 individuals\n Min 76.0\n Max 89.0\n Avg 85.1\n Std 2.20529665427\n-- Generation 19 --\n Evaluated 189 individuals\n Min 75.0\n Max 90.0\n Avg 85.77\n Std 2.1564863397\n-- Generation 20 --\n Evaluated 188 individuals\n Min 77.0\n Max 91.0\n Avg 86.4833333333\n Std 2.2589943682\n-- Generation 21 --\n Evaluated 180 individuals\n Min 80.0\n Max 91.0\n Avg 87.24\n Std 2.0613264338\n-- Generation 22 --\n Evaluated 179 individuals\n Min 80.0\n Max 92.0\n Avg 87.95\n Std 1.95298916194\n-- Generation 23 --\n Evaluated 196 individuals\n Min 79.0\n Max 93.0\n Avg 88.42\n Std 2.2249194742\n-- Generation 24 --\n Evaluated 168 individuals\n Min 82.0\n Max 93.0\n Avg 89.2833333333\n Std 1.89289607627\n-- Generation 25 --\n Evaluated 186 individuals\n Min 78.0\n Max 94.0\n Avg 89.7666666667\n Std 2.26102238428\n-- Generation 26 --\n Evaluated 182 individuals\n Min 82.0\n Max 94.0\n Avg 90.4633333333\n Std 2.21404356075\n-- Generation 27 --\n Evaluated 179 individuals\n Min 81.0\n Max 95.0\n Avg 90.8733333333\n Std 2.41328729238\n-- Generation 28 --\n Evaluated 183 individuals\n Min 83.0\n Max 95.0\n Avg 91.7166666667\n Std 2.18701978856\n-- Generation 29 --\n Evaluated 167 individuals\n Min 83.0\n Max 98.0\n Avg 92.3466666667\n Std 2.21656390739\n-- Generation 30 --\n Evaluated 170 individuals\n Min 84.0\n Max 98.0\n Avg 92.9533333333\n Std 2.09868742048\n-- Generation 31 --\n Evaluated 172 individuals\n Min 83.0\n Max 97.0\n Avg 93.5266666667\n Std 2.28238666507\n-- Generation 32 --\n Evaluated 196 individuals\n Min 86.0\n Max 98.0\n Avg 94.28\n Std 2.16985406575\n-- Generation 33 --\n Evaluated 176 individuals\n Min 85.0\n Max 98.0\n Avg 94.9133333333\n Std 2.22392046221\n-- Generation 34 --\n Evaluated 176 individuals\n Min 86.0\n Max 99.0\n Avg 95.6333333333\n Std 2.13359373411\n-- Generation 35 --\n Evaluated 174 individuals\n Min 86.0\n Max 99.0\n Avg 96.2966666667\n Std 2.23651266236\n-- Generation 36 --\n Evaluated 174 individuals\n Min 87.0\n Max 100.0\n Avg 96.5866666667\n Std 2.41436442062\n-- Generation 37 --\n Evaluated 195 individuals\n Min 84.0\n Max 100.0\n Avg 97.3666666667\n Std 2.16153237825\n-- Generation 38 --\n Evaluated 180 individuals\n Min 89.0\n Max 100.0\n Avg 97.7466666667\n Std 2.32719191779\n-- Generation 39 --\n Evaluated 196 individuals\n Min 88.0\n Max 100.0\n Avg 98.1833333333\n Std 2.33589145486\n"
],
[
"best_ind = tools.selBest(pop, 1)[0]\nprint(\"Best individual is %s, %s\" % (best_ind, best_ind.fitness.values))",
"Best individual is [1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1], (100.0,)\n"
]
],
[
[
"## Symbolic Regression (GP)",
"_____no_output_____"
],
[
"Symbolic regression is one of the best known problems in GP. It is commonly used as a tuning problem for new algorithms, but is also widely used with real-life distributions, where other regression methods may not work.\n\nAll symbolic regression problems use an arbitrary data distribution, and try to fit the most accurately the data with a symbolic formula. Usually, a measure like the RMSE (Root Mean Square Error) is used to measure an individual’s fitness.\n\nIn this example, we use a classical distribution, the quartic polynomial (x^4 + x^3 + x^2 + x), a one-dimension distribution. 20 equidistant points are generated in the range [-1, 1], and are used to evaluate the fitness.",
"_____no_output_____"
]
],
[
[
"import operator\nimport math\nimport random\n\nimport numpy\n\nfrom deap import algorithms\nfrom deap import base\nfrom deap import creator\nfrom deap import tools\nfrom deap import gp\n\n# define a new function for divison that guards against divide by 0\ndef protectedDiv(left, right):\n try:\n return left / right\n except ZeroDivisionError:\n return 1",
"_____no_output_____"
],
[
"# add aritmetic primitives\npset = gp.PrimitiveSet(\"MAIN\", 1)\npset.addPrimitive(operator.add, 2)\npset.addPrimitive(operator.sub, 2)\npset.addPrimitive(operator.mul, 2)\npset.addPrimitive(protectedDiv, 2)\npset.addPrimitive(operator.neg, 1)\npset.addPrimitive(math.cos, 1)\npset.addPrimitive(math.sin, 1)\n\n# constant terminal\npset.addEphemeralConstant(\"rand101\", lambda: random.randint(-1,1))\n\n# define number of inputs\npset.renameArguments(ARG0='x')",
"_____no_output_____"
],
[
"# create fitness and individual objects\ncreator.create(\"FitnessMin\", base.Fitness, weights=(-1.0,))\ncreator.create(\"Individual\", gp.PrimitiveTree, fitness=creator.FitnessMin)",
"_____no_output_____"
],
[
"# register evolution process parameters through the toolbox\ntoolbox = base.Toolbox()\ntoolbox.register(\"expr\", gp.genHalfAndHalf, pset=pset, min_=1, max_=2)\ntoolbox.register(\"individual\", tools.initIterate, creator.Individual, toolbox.expr)\ntoolbox.register(\"population\", tools.initRepeat, list, toolbox.individual)\ntoolbox.register(\"compile\", gp.compile, pset=pset)\n\n# evaluation function\ndef evalSymbReg(individual, points):\n # transform the tree expression in a callable function\n func = toolbox.compile(expr=individual)\n # evaluate the mean squared error between the expression\n # and the real function : x**4 + x**3 + x**2 + x\n sqerrors = ((func(x) - x**4 - x**3 - x**2 - x)**2 for x in points)\n return math.fsum(sqerrors) / len(points),\n\ntoolbox.register(\"evaluate\", evalSymbReg, points=[x/10. for x in range(-10,10)])\ntoolbox.register(\"select\", tools.selTournament, tournsize=3)\ntoolbox.register(\"mate\", gp.cxOnePoint)\ntoolbox.register(\"expr_mut\", gp.genFull, min_=0, max_=2)\ntoolbox.register(\"mutate\", gp.mutUniform, expr=toolbox.expr_mut, pset=pset)\n\n# prevent functions from getting too deep/complex\ntoolbox.decorate(\"mate\", gp.staticLimit(key=operator.attrgetter(\"height\"), max_value=17))\ntoolbox.decorate(\"mutate\", gp.staticLimit(key=operator.attrgetter(\"height\"), max_value=17))",
"_____no_output_____"
],
[
"# compute some statistics about the population\nstats_fit = tools.Statistics(lambda ind: ind.fitness.values)\nstats_size = tools.Statistics(len)\nmstats = tools.MultiStatistics(fitness=stats_fit, size=stats_size)\nmstats.register(\"avg\", numpy.mean)\nmstats.register(\"std\", numpy.std)\nmstats.register(\"min\", numpy.min)\nmstats.register(\"max\", numpy.max)",
"_____no_output_____"
],
[
"random.seed(318)\n\npop = toolbox.population(n=300)\nhof = tools.HallOfFame(1)\n\n# run the algorithm\npop, log = algorithms.eaSimple(pop, toolbox, 0.5, 0.1, 40, stats=mstats,\n halloffame=hof, verbose=True)",
" \t \t fitness \t size \n \t \t---------------------------------------\t-------------------------------\ngen\tnevals\tavg \tmax \tmin \tstd \tavg \tmax\tmin\tstd \n0 \t300 \t2.39949\t59.2593\t0.165572\t4.64122\t3.69667\t7 \t2 \t1.61389\n1 \t146 \t1.0971 \t10.1 \t0.165572\t0.845978\t3.80667\t13 \t1 \t1.78586\n2 \t169 \t0.902365\t6.5179 \t0.165572\t0.72362 \t4.16 \t13 \t1 \t2.0366 \n3 \t167 \t0.852725\t9.6327 \t0.165572\t0.869381\t4.63667\t13 \t1 \t2.20408\n4 \t158 \t0.74829 \t14.1573\t0.165572\t1.01281 \t4.88333\t13 \t1 \t2.14392\n5 \t160 \t0.630299\t7.90605\t0.165572\t0.904373\t5.52333\t14 \t1 \t2.09351\n6 \t181 \t0.495118\t4.09456\t0.165572\t0.524658\t6.08333\t13 \t1 \t1.99409\n7 \t170 \t0.403873\t2.6434 \t0.165572\t0.440596\t6.34667\t14 \t1 \t1.84386\n8 \t173 \t0.393405\t2.9829 \t0.165572\t0.425415\t6.37 \t12 \t1 \t1.78132\n9 \t168 \t0.414299\t13.5996\t0.165572\t0.841226\t6.25333\t11 \t2 \t1.76328\n10 \t142 \t0.384179\t4.07808\t0.165572\t0.477269\t6.25667\t13 \t1 \t1.78067\n11 \t156 \t0.459639\t19.8316\t0.165572\t1.47254 \t6.35333\t15 \t1 \t2.04983\n12 \t167 \t0.384348\t6.79674\t0.165572\t0.495807\t6.25 \t13 \t1 \t1.92029\n13 \t157 \t0.42446 \t11.0636\t0.165572\t0.818953\t6.43667\t15 \t1 \t2.11959\n14 \t175 \t0.342257\t2.552 \t0.165572\t0.325872\t6.23333\t15 \t1 \t2.14295\n15 \t154 \t0.442374\t13.8349\t0.165572\t0.950612\t6.05667\t14 \t1 \t1.90266\n16 \t181 \t0.455697\t19.7228\t0.101561\t1.39528 \t6.08667\t13 \t1 \t1.84006\n17 \t178 \t0.36256 \t2.54124\t0.101561\t0.340555\t6.24 \t15 \t1 \t2.20055\n18 \t171 \t0.411532\t14.2339\t0.101561\t0.897785\t6.44 \t15 \t1 \t2.2715 \n19 \t156 \t0.43193 \t15.5923\t0.101561\t0.9949 \t6.66667\t15 \t1 \t2.40185\n20 \t169 \t0.398163\t4.09456\t0.0976781\t0.450231\t6.96667\t15 \t1 \t2.62022\n21 \t162 \t0.385774\t4.09456\t0.0976781\t0.421867\t7.13 \t14 \t1 \t2.65577\n22 \t162 \t0.35318 \t2.55465\t0.0253803\t0.389453\t7.66667\t19 \t2 \t3.04995\n23 \t164 \t0.3471 \t3.66792\t0.0253803\t0.482334\t8.24 \t21 \t1 \t3.48364\n24 \t159 \t1.46248 \t331.247\t0.0253803\t19.0841 \t9.42667\t19 \t3 \t3.238 \n25 \t164 \t0.382697\t6.6452 \t0.0173316\t0.652247\t10.1867\t25 \t1 \t3.46292\n26 \t139 \t0.367651\t11.9045\t0.0173316\t0.855067\t10.67 \t19 \t3 \t3.32582\n27 \t167 \t0.345866\t6.6452 \t0.0173316\t0.586155\t11.4 \t27 \t3 \t3.44384\n28 \t183 \t0.388404\t4.53076\t0.0173316\t0.58986 \t11.5767\t24 \t3 \t3.4483 \n29 \t163 \t0.356009\t6.33264\t0.0173316\t0.563266\t12.2433\t29 \t2 \t4.23211\n30 \t174 \t0.31506 \t2.54124\t0.0173316\t0.412507\t12.92 \t27 \t3 \t4.5041 \n31 \t206 \t0.361197\t2.9829 \t0.0173316\t0.486155\t13.9333\t33 \t1 \t5.6747 \n32 \t168 \t0.302704\t4.01244\t0.0173316\t0.502277\t15.04 \t31 \t3 \t5.40849\n33 \t160 \t0.246509\t3.30873\t0.012947 \t0.433212\t16.3967\t34 \t2 \t5.66092\n34 \t158 \t0.344791\t26.1966\t0.012947 \t1.57277 \t17.39 \t43 \t1 \t6.13008\n35 \t162 \t0.212572\t2.85856\t0.0148373\t0.363023\t17.64 \t37 \t2 \t6.04349\n36 \t183 \t0.240268\t5.06093\t0.0112887\t0.482794\t17.4333\t41 \t3 \t6.33184\n37 \t185 \t0.514635\t65.543 \t0.0103125\t3.7864 \t16.6167\t41 \t1 \t6.58456\n38 \t134 \t0.340433\t11.2506\t0.0103125\t0.827213\t16.2733\t34 \t1 \t6.08484\n39 \t158 \t0.329797\t15.8145\t4.50668e-33\t1.05693 \t16.4133\t34 \t1 \t6.09993\n40 \t164 \t0.306543\t14.3573\t4.50668e-33\t0.947046\t17.9033\t53 \t2 \t8.23695\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b5bd024f4ace1ec9343fb4334ffbf506d28622 | 130,521 | ipynb | Jupyter Notebook | pure-text/preprocessing-pdf.ipynb | huseinzol05/Malay-Dataset | e27b7617c74395c86bb5ed9f3f194b3cac2f66f6 | [
"Apache-2.0"
] | 51 | 2020-05-20T13:26:18.000Z | 2021-05-13T07:21:17.000Z | pure-text/preprocessing-pdf.ipynb | huseinzol05/Malay-Dataset | e27b7617c74395c86bb5ed9f3f194b3cac2f66f6 | [
"Apache-2.0"
] | 3 | 2020-05-21T13:12:46.000Z | 2021-05-12T03:26:43.000Z | pure-text/preprocessing-pdf.ipynb | huseinzol05/Malaya-Dataset | c9c1917a6b1cab823aef5a73bd10e0fab0bff42d | [
"Apache-2.0"
] | 21 | 2019-02-08T05:17:24.000Z | 2020-05-05T09:28:50.000Z | 72.310803 | 702 | 0.668812 | [
[
[
"# !wget https://malaya-dataset.s3-ap-southeast-1.amazonaws.com/crawler/academia/academia-pdf.json",
"_____no_output_____"
],
[
"import json\nimport cleaning\nfrom tqdm import tqdm",
"_____no_output_____"
],
[
"with open('../academia/academia-pdf.json') as fopen:\n pdf = json.load(fopen)\n \nlen(pdf)",
"_____no_output_____"
],
[
"import os\n\nos.path.split(pdf[0]['file'])",
"_____no_output_____"
],
[
"import malaya\n\nfast_text = malaya.language_detection.fasttext()",
"\n"
],
[
"fast_text.predict(['Prosiding_Kolokium_Siswazah_JUF_2017.pdf'])",
"_____no_output_____"
],
[
"from unidecode import unidecode\n\ndef clean(string):\n string = [cleaning.cleaning(s) for s in string]\n \n string = [s.strip() for s in string if 'tarikh' not in s.lower() and 'soalan no' not in s.lower()]\n string = [s for s in string if not ''.join(s.split()[:1]).isdigit() and '.soalan' not in s.lower() and 'jum ' not in s.lower()]\n string = [s for s in string if not s[:3].isdigit() and not s[-3:].isdigit()]\n return string",
"_____no_output_____"
],
[
"outer = []\n\nfor k in tqdm(range(len(pdf))):\n\n c = clean(pdf[k]['content']['content'].split('\\n'))\n t, last = [], 0\n\n i = 0\n while i < len(c):\n text = c[i]\n\n if len(text) > 5:\n if len(text.split()) > 1:\n t.append(text)\n last = i\n else:\n if len(t) and (i - last) > 2:\n t.append('')\n outer.extend(t)\n t = []\n last = i\n elif not len(t):\n last = i\n\n i += 1\n \n if len(t):\n t.append('')\n outer.extend(t)",
"100%|██████████| 1414/1414 [07:04<00:00, 3.33it/s]\n"
],
[
"len(outer)",
"_____no_output_____"
],
[
"%%time\n\ntemp_vocab = list(set(cleaning.multiprocessing(outer, cleaning.unique_words)))",
"CPU times: user 3.93 s, sys: 2.22 s, total: 6.15 s\nWall time: 7.15 s\n"
],
[
"%%time\n\n# important\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.duplicate_dots_marks_exclamations, list_mode = False)\nprint(len(temp_dict))",
"7040\nCPU times: user 415 ms, sys: 961 ms, total: 1.38 s\nWall time: 3.11 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:32<00:00, 276346.68it/s]\n"
],
[
"%%time\n\n# important\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_underscore, list_mode = False)\nprint(len(temp_dict))",
"536\nCPU times: user 591 ms, sys: 972 ms, total: 1.56 s\nWall time: 2.19 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:32<00:00, 274032.82it/s]\n"
],
[
"%%time\n\n# important\ntemp_dict = cleaning.multiprocessing(outer, cleaning.isolate_spamchars, list_mode = False)\nprint(len(temp_dict))",
"0\nCPU times: user 2.54 s, sys: 2.15 s, total: 4.69 s\nWall time: 10.7 s\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.break_short_words, list_mode = False)\nprint(len(temp_dict))",
"19693\nCPU times: user 407 ms, sys: 1.01 s, total: 1.42 s\nWall time: 1.55 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:32<00:00, 270720.18it/s]\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.break_long_words, list_mode = False)\nprint(len(temp_dict))",
"4441\nCPU times: user 360 ms, sys: 1.04 s, total: 1.4 s\nWall time: 1.64 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:32<00:00, 278113.08it/s]\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_ending_underscore, list_mode = False)\nprint(len(temp_dict))",
"272\nCPU times: user 314 ms, sys: 1 s, total: 1.31 s\nWall time: 1.5 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:32<00:00, 270761.52it/s]\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.remove_starting_underscore, list_mode = False)\nprint(len(temp_dict))",
"343\nCPU times: user 376 ms, sys: 1 s, total: 1.38 s\nWall time: 1.57 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:32<00:00, 278484.09it/s]\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.end_punct, list_mode = False)\nprint(len(temp_dict))",
"533165\nCPU times: user 2.05 s, sys: 1.08 s, total: 3.13 s\nWall time: 3.32 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:35<00:00, 249997.35it/s]\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.start_punct, list_mode = False)\nprint(len(temp_dict))",
"178877\nCPU times: user 949 ms, sys: 1.04 s, total: 1.99 s\nWall time: 2.16 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:35<00:00, 249939.42it/s]\n"
],
[
"%%time\ntemp_dict = cleaning.multiprocessing(temp_vocab, cleaning.join_dashes, list_mode = False)\nprint(len(temp_dict))",
"495\nCPU times: user 319 ms, sys: 1 s, total: 1.32 s\nWall time: 1.63 s\n"
],
[
"outer = cleaning.string_dict_cleaning(outer, temp_dict)",
"100%|██████████| 8926305/8926305 [00:35<00:00, 253898.14it/s]\n"
],
[
"results, result = [], []\nfor i in tqdm(outer):\n if not len(i) and len(result):\n results.append(result)\n result = []\n else:\n result.append(i)\n \nif len(result):\n results.append(result)",
"100%|██████████| 8926305/8926305 [00:07<00:00, 1261203.88it/s]\n"
],
[
"import re\n\nalphabets = '([A-Za-z])'\nprefixes = (\n '(Mr|St|Mrs|Ms|Dr|Prof|Capt|Cpt|Lt|Mt|Puan|puan|Tuan|tuan|sir|Sir)[.]'\n)\nsuffixes = '(Inc|Ltd|Jr|Sr|Co|Mo)'\nstarters = '(Mr|Mrs|Ms|Dr|He\\s|She\\s|It\\s|They\\s|Their\\s|Our\\s|We\\s|But\\s|However\\s|That\\s|This\\s|Wherever|Dia|Mereka|Tetapi|Kita|Itu|Ini|Dan|Kami|Beliau|Seri|Datuk|Dato|Datin|Tuan|Puan)'\nacronyms = '([A-Z][.][A-Z][.](?:[A-Z][.])?)'\nwebsites = '[.](com|net|org|io|gov|me|edu|my)'\nanother_websites = '(www|http|https)[.]'\ndigits = '([0-9])'\nbefore_digits = '([Nn]o|[Nn]ombor|[Nn]umber|[Kk]e|=|al)'\nmonth = '([Jj]an(?:uari)?|[Ff]eb(?:ruari)?|[Mm]a(?:c)?|[Aa]pr(?:il)?|Mei|[Jj]u(?:n)?|[Jj]ula(?:i)?|[Aa]ug(?:ust)?|[Ss]ept?(?:ember)?|[Oo]kt(?:ober)?|[Nn]ov(?:ember)?|[Dd]is(?:ember)?)'\n\n\ndef split_into_sentences(text, minimum_length = 5):\n text = text.replace('\\x97', '\\n')\n text = '. '.join([s for s in text.split('\\n') if len(s)])\n text = text + '.'\n text = unidecode(text)\n text = ' ' + text + ' '\n text = text.replace('\\n', ' ')\n text = re.sub(prefixes, '\\\\1<prd>', text)\n text = re.sub(websites, '<prd>\\\\1', text)\n text = re.sub(another_websites, '\\\\1<prd>', text)\n text = re.sub('[,][.]+', '<prd>', text)\n if '...' in text:\n text = text.replace('...', '<prd><prd><prd>')\n if 'Ph.D' in text:\n text = text.replace('Ph.D.', 'Ph<prd>D<prd>')\n text = re.sub('[.]\\s*[,]', '<prd>,', text)\n text = re.sub(before_digits + '\\s*[.]\\s*' + digits, '\\\\1<prd>\\\\2', text)\n text = re.sub(month + '[.]\\s*' + digits, '\\\\1<prd>\\\\2', text)\n text = re.sub('\\s' + alphabets + '[.][ ]+', ' \\\\1<prd> ', text)\n text = re.sub(acronyms + ' ' + starters, '\\\\1<stop> \\\\2', text)\n text = re.sub(\n alphabets + '[.]' + alphabets + '[.]' + alphabets + '[.]',\n '\\\\1<prd>\\\\2<prd>\\\\3<prd>',\n text,\n )\n text = re.sub(\n alphabets + '[.]' + alphabets + '[.]', '\\\\1<prd>\\\\2<prd>', text\n )\n text = re.sub(' ' + suffixes + '[.][ ]+' + starters, ' \\\\1<stop> \\\\2', text)\n text = re.sub(' ' + suffixes + '[.]', ' \\\\1<prd>', text)\n text = re.sub(' ' + alphabets + '[.]', ' \\\\1<prd>', text)\n text = re.sub(digits + '[.]' + digits, '\\\\1<prd>\\\\2', text)\n if '”' in text:\n text = text.replace('.”', '”.')\n if '\"' in text:\n text = text.replace('.\"', '\".')\n if '!' in text:\n text = text.replace('!\"', '\"!')\n if '?' in text:\n text = text.replace('?\"', '\"?')\n text = text.replace('.', '.<stop>')\n text = text.replace('?', '?<stop>')\n text = text.replace('!', '!<stop>')\n text = text.replace('<prd>', '.')\n sentences = text.split('<stop>')\n sentences = sentences[:-1]\n sentences = [s.strip() for s in sentences if len(s) > minimum_length]\n return sentences\n\nsplit_into_sentences('733 ke . 633 , berlaku penurunan akibat kesan program PMI .')",
"_____no_output_____"
],
[
"import malaya\nimport re\n\ndef strip(string):\n string = ' '.join(string)\n string = re.sub(r'[ ]+', ' ', string.replace('\\n', ' ').replace('\\t', ' ')).strip()\n return split_into_sentences(string)",
"_____no_output_____"
],
[
"output = []\n\nfor r in tqdm(results):\n output.extend(strip(r) + [''])",
"100%|██████████| 678289/678289 [04:47<00:00, 2360.23it/s]\n"
],
[
"len(output)",
"_____no_output_____"
],
[
"output[10000:11000]",
"_____no_output_____"
],
[
"with open('dumping-academia.txt', 'w') as fopen:\n fopen.write('\\n'.join(output))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b5d75c7aee2386a2df304ff43088874c8daf0a | 3,329 | ipynb | Jupyter Notebook | 0_Bienvenida/examen_diagnostico.ipynb | nextia-academy/analisis-datos-python | a0dfc87dd0620ef5e37ea972ed25538b9c151b63 | [
"MIT"
] | 1 | 2020-05-12T20:26:46.000Z | 2020-05-12T20:26:46.000Z | 0_Bienvenida/examen_diagnostico.ipynb | pazcuellar/analisis-datos-python | a12e8104ee579a5194391884c169f950b51164a0 | [
"MIT"
] | null | null | null | 0_Bienvenida/examen_diagnostico.ipynb | pazcuellar/analisis-datos-python | a12e8104ee579a5194391884c169f950b51164a0 | [
"MIT"
] | null | null | null | 29.460177 | 180 | 0.487233 | [
[
[
"# Examen diagnóstico\n\n---\n\n**Instrucciones: contesta el siguiente examen en el lenguaje de programación de tu preferencia, o bien, en pseudocódigo. Envía tus resspuetas al correo [email protected]**\n\n$\\text{1. Escribe un programa que reciba la base y altura de un tríangulo y devuelva el área de éste. Recuerda que}$\n$$\nA = \\frac{base \\times altura}{2}\n$$\n```\n base = 10\n altura = 5\n >> 25\n```\n---\n$\\text{2. Dado un número entero } n \\text{ , imprime si éste es par o impar}$\n```\n n = 4\n >> \"El número es par\"\n```\n---\n$\\text{3. Dado el siguiente arreglo:}$\n\n`arr = [14,62,23,52,87,33,72]` \n\n$\\text{¿cuáles son los elementos que se encuentran en los índices 1, 3 y 5?}$\n\n---\n$\\text{4. Escribe un programa que imprima los primeros 15 elementos de la serie de Fibonacci,}$\n```\n >> 0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, 233, 377\n```\n---\n$\\text{5. Escribe un programa que imprima la suma de los primeros cien números naturales (1 y 100 incluídos)}$\n```\n >> 5050\n```\n---\n$\\text{6. Dado un número entero } n \\text{ , y una listas de números, imprime si el número aparece en la lista}$\n```\n lista = [5, 4, 9, 100, 29, 143, 2, 456, 2, 201, 34, 49, 0]\n n = 9\n >> \"Sí está en la lista\"\n```\n---\n$\\text{7. Dado un string, cuenta e imprime la frecuencia con la que aparece la letra 'a' en dicho string}$\n```\n str = \"ciencia de datos con Gerardo\"\n >> 3\n```\n---\n$\\text{8. Escribe un programa que reciba el diámetro de un círculo e imprima el área y la circunferencia de éste. Recuerda que}$\n$$\nA = \\pi r^2 \\hspace{1cm} \\text{donde } r \\rightarrow radio\n$$\n```\n diametro = 10\n >> 78.54\n >> 31.42\n```\n---\n$\\text{9. Escribe un programa que reciba el nombre del usuario y posteriormente imprima un saludo }$\n```\n nombre = 'Gerardo'\n >> \"Hola Gerardo, bienvenido al curso\"\n```\n---\n$\\text{10. Escribe un programa que reciba la edad del usuario e imprima si puede o no votar.}$\n```\n edad = 25\n >> \"Sí puede votar\"\n```\n```\n edad = 17\n >> \"No puede votar\"\n```\n```\n edad = 18\n >> \"Sí puede votar\"\n```",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown"
]
] |
d0b5dc5072da61110340cd18c068bd78f11f8251 | 77,165 | ipynb | Jupyter Notebook | imersao_dados_aula_1.ipynb | CarlosNeto2804/imersao-dados-2 | cff8df93e8ca2c241f48e475756c72d66046567a | [
"MIT"
] | null | null | null | imersao_dados_aula_1.ipynb | CarlosNeto2804/imersao-dados-2 | cff8df93e8ca2c241f48e475756c72d66046567a | [
"MIT"
] | null | null | null | imersao_dados_aula_1.ipynb | CarlosNeto2804/imersao-dados-2 | cff8df93e8ca2c241f48e475756c72d66046567a | [
"MIT"
] | null | null | null | 95.030788 | 13,786 | 0.773913 | [
[
[
"<a href=\"https://colab.research.google.com/github/CarlosNeto2804/imersao-dados-2/blob/main/imersao_dados_aula_1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Introdução",
"_____no_output_____"
]
],
[
[
"import pandas as pd;\n\ndados_enem = pd.read_csv('https://github.com/alura-cursos/imersao-dados-2-2020/blob/master/MICRODADOS_ENEM_2019_SAMPLE_43278.csv?raw=true')\n\n# retorna as 5 primeiras linhas da colecao\ndados_enem.head() \n\n# para acessar apenas uma coluna -> dados_enem['nome_da_coluna']\ndados_enem['SG_UF_RESIDENCIA']\n\n# para verificar quais colunas existem no data frame -> dados_enem.colums.values\ndados_enem.columns.values\n\n# acessar mais de um valor no DataFrame data_frame[[\"cabeçalho1\",\"cabeçalho2\"]]\ndados_enem[[\"SG_UF_RESIDENCIA\",\"Q025\"]]\n\n# retornar os valores sem repeticao de uma coluna\ndados_enem[\"SG_UF_RESIDENCIA\"].unique() ",
"_____no_output_____"
],
[
"# soma dos elementos de uma derminada chave\n# ordena pelo valor\n#ESTADOS\ndados_enem[\"SG_UF_RESIDENCIA\"].value_counts()\n\n#IDADES\ndados_enem['NU_IDADE'].value_counts()\n\n# ordenacao pelo index\ndados_enem['NU_IDADE'].value_counts().sort_index()\ndados_enem['NU_IDADE'].describe()\n",
"_____no_output_____"
]
],
[
[
"# Continuação",
"_____no_output_____"
]
],
[
[
"# visualização em histograma\n#dados_enem['NU_IDADE'].hist()\ndados_enem['NU_IDADE'].hist(bins=100,figsize=(11,9),legend=True)",
"_____no_output_____"
],
[
"treineiros = dados_enem.query('IN_TREINEIRO == 1')\ntreineiros['NU_IDADE'].value_counts()",
"_____no_output_____"
],
[
"# Notas Redacao\ndados_enem['NU_NOTA_REDACAO'].hist(bins=20)",
"_____no_output_____"
],
[
"# analise geral\nprovas = [\"NU_NOTA_CN\",\"NU_NOTA_CH\",\"NU_NOTA_MT\",\"NU_NOTA_LC\",\"NU_NOTA_REDACAO\"]\ndados_enem[provas].describe()",
"_____no_output_____"
]
],
[
[
"# Desafios\n",
"_____no_output_____"
],
[
"- 01 : Informar a proporção de inscritos por idades",
"_____no_output_____"
]
],
[
[
"# desafio 1\ndef proporcao(total_itens):\n def funcao_calculo(x): \n res = x * 100 / total_itens\n return round(res, 6);\n return funcao_calculo\n\ntotal = len(dados_enem)\ninscritos_por_idade = dados_enem['NU_IDADE'].value_counts()\ninscritos_por_idade.apply(proporcao(total))",
"_____no_output_____"
]
],
[
[
"- 02: Descobrir de quais estados são os inscritos com 13 anos",
"_____no_output_____"
]
],
[
[
"# desafio 02\ncabecalhos= ['SG_UF_RESIDENCIA','NU_IDADE']\ninscritos=dados_enem[cabecalhos]\ninscritos.query('NU_IDADE==13')\n",
"_____no_output_____"
]
],
[
[
"- 03: Qual a proporcao dos alunos com 18 anos por estado",
"_____no_output_____"
]
],
[
[
"# desafio 3\ncabecalhos = ['SG_UF_RESIDENCIA','NU_IDADE']\nnovo_df = dados_enem[cabecalhos]\ninscritos = novo_df.query('NU_IDADE==18')\ntotal_inscritos = len(inscritos)\ninscritos.value_counts().apply(proporcao(total_inscritos))\n",
"_____no_output_____"
]
],
[
[
"- 04: Plotar Histogramas das idades de treineiros e não treineiros",
"_____no_output_____"
]
],
[
[
"# desafio 4\ninscritos_treineiros = dados_enem.query('IN_TREINEIRO == 1')['NU_IDADE'].value_counts();\ninscritos_treineiros.hist(bins=30,figsize=(10,7),legend=True)",
"_____no_output_____"
],
[
"inscritos_nao_treineiros = dados_enem.query('IN_TREINEIRO == 0')['NU_IDADE'].value_counts();\ninscritos_nao_treineiros.hist(bins=30,figsize=(10,7),legend=True)",
"_____no_output_____"
]
],
[
[
"- 05: Comparar as distribuições das provas em ingles e espanhol",
"_____no_output_____"
]
],
[
[
"# desafio 5\n# TP_LINGUA==0 -> Ingles\n# TP_LINGUA==1 -> Espanhol\ndados_enem.query('TP_LINGUA==1')['TP_LINGUA'].hist(bins=20,figsize=(10,7),legend=True)\ndados_enem.query('TP_LINGUA==0')['TP_LINGUA'].hist(bins=20,figsize=(10,7),legend=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b5e4fd15b39e9a2b9901381a4e874e55f7aabb | 5,885 | ipynb | Jupyter Notebook | nb/singleTable_example.ipynb | LSSTDESC/tables_io | 1c2f119c928d05d237b1c8509e340d29650ceb8b | [
"MIT"
] | 1 | 2021-08-13T15:41:58.000Z | 2021-08-13T15:41:58.000Z | nb/singleTable_example.ipynb | LSSTDESC/tables_io | 1c2f119c928d05d237b1c8509e340d29650ceb8b | [
"MIT"
] | 18 | 2021-08-12T00:09:36.000Z | 2022-02-24T21:11:18.000Z | nb/singleTable_example.ipynb | LSSTDESC/tables_io | 1c2f119c928d05d237b1c8509e340d29650ceb8b | [
"MIT"
] | null | null | null | 23.634538 | 162 | 0.549873 | [
[
[
"# Using `tables_io.read`, `tables_io.write` and `tables_io.convert`\n\nThese functions can be used to read and write single tables and to convert them to different formats\n\nThe Tables can be in any of the formats that `tables_io` supports, see more on that in the notebook below.\n\nLet's have a look",
"_____no_output_____"
]
],
[
[
"# Standard imports\nimport os\nimport numpy as np\nimport tables_io\nfrom tables_io.testUtils import make_test_data",
"_____no_output_____"
],
[
"# make several tables and grab one\ntables = make_test_data()\ndata = tables['data']\ndata",
"_____no_output_____"
],
[
"data_np = tables_io.convert(data, tables_io.types.NUMPY_DICT)\ndata_np",
"_____no_output_____"
],
[
"data_pd = tables_io.convert(data, tables_io.types.PD_DATAFRAME)\ndata_pd",
"_____no_output_____"
]
],
[
[
"# File IO with `tables_io`\n\nWe can write tables into several different formats. These include:\n\n1. fits: Writing `astropy.table.Table` objects to FITS files (with the suffix 'fits')\n2. hf5: Writing `astropy.table.Table` objects to HDF5 files (with the suffix 'hf5')\n3. hfd5: Writing `numpy.array` objects to HDF5 files (with the suffix 'hdf5')\n4. h5: Writing `pandas.DataFrame` objects to HDF5 files (with the suffix 'h5')\n5. pq: Writing `pandas.DataFrame` objects to parquet files (with the suffix 'pq')\n\nAlso, each table type has a 'native' format that we use as a default. Setting the `fmt` to `None` in function calls will typically use the 'native' format.",
"_____no_output_____"
]
],
[
[
"all_fmts = list(tables_io.types.FILE_FORMAT_SUFFIXS.keys()) + [None]\nprint(all_fmts)",
"_____no_output_____"
]
],
[
[
"# Ok let's write the data to different files",
"_____no_output_____"
]
],
[
[
"for fmt in all_fmts:\n if fmt is None:\n basename = 'test_single_native'\n else:\n basename = 'test_single_out'\n print(\"Writing to %s using format %s\" % (basename, fmt))\n try:\n os.unlink('%s.%s' % (basename, fmt))\n except:\n pass\n try:\n tables_io.write(data, basename, fmt)\n except ImportError as msg:\n print(\"Skipping format %s because %s\" % (fmt, msg))",
"_____no_output_____"
],
[
"! ls test_single_*",
"_____no_output_____"
]
],
[
[
"# Ok, now let's read things back",
"_____no_output_____"
]
],
[
[
"data_r_fits = tables_io.read(\"test_single_out.fits\")\ndata_r_fits",
"_____no_output_____"
],
[
"data_r_hdf5 = tables_io.read(\"test_single_out.hdf5\")\ndata_r_hdf5",
"_____no_output_____"
],
[
"data_r_hf5 = tables_io.read(\"test_single_out.hf5\")\ndata_r_hf5",
"_____no_output_____"
],
[
"data_r_pq = tables_io.read(\"test_single_out.pq\", keys=[''])\ndata_r_pq",
"_____no_output_____"
],
[
"data_r_h5 = tables_io.read(\"test_single_out.h5\")\ndata_r_h5",
"_____no_output_____"
],
[
"data_native = tables_io.read(\"test_single_native.hf5\")\ndata_native",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b5e86f7ad11526a92f778a88366184fb2db9cc | 2,331 | ipynb | Jupyter Notebook | Iris Flower Assignment/Iris Flower Dataset Assignment.ipynb | teenage-coder/Data-Science-with-Python | 436510b649dfbfc0bcfed951063dea0606afe45e | [
"Apache-2.0"
] | 1 | 2021-09-16T03:17:18.000Z | 2021-09-16T03:17:18.000Z | Iris Flower Assignment/Iris Flower Dataset Assignment.ipynb | teenage-coder/Data-Science-with-Python | 436510b649dfbfc0bcfed951063dea0606afe45e | [
"Apache-2.0"
] | null | null | null | Iris Flower Assignment/Iris Flower Dataset Assignment.ipynb | teenage-coder/Data-Science-with-Python | 436510b649dfbfc0bcfed951063dea0606afe45e | [
"Apache-2.0"
] | null | null | null | 16.891304 | 68 | 0.493779 | [
[
[
"# Q1. How many types of flowers we are having in this Dataset?",
"_____no_output_____"
],
[
"# Q2. Frequency of Every Category Flower",
"_____no_output_____"
],
[
"# Q3. Plot a bar graph on Flower Category Frequency",
"_____no_output_____"
],
[
"# Q4. Preprocess the Features and Labels",
"_____no_output_____"
],
[
"# Q5. Train Test and Validation Split (80/10/10)",
"_____no_output_____"
],
[
"# Q6. Build a ANN based Model",
"_____no_output_____"
],
[
"# Q7. Train the Model",
"_____no_output_____"
],
[
"# Q8. Model Evaluation",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0b5f1ae140e156da4111dd030eabc95c3ce1374 | 19,963 | ipynb | Jupyter Notebook | examples/.ipynb_checkpoints/example_Polygons_reduction-checkpoint.ipynb | PhilipeRLeal/time_space_reductions | 8d826d0541846fac4e869db88c0b55794b32752f | [
"MIT"
] | null | null | null | examples/.ipynb_checkpoints/example_Polygons_reduction-checkpoint.ipynb | PhilipeRLeal/time_space_reductions | 8d826d0541846fac4e869db88c0b55794b32752f | [
"MIT"
] | null | null | null | examples/.ipynb_checkpoints/example_Polygons_reduction-checkpoint.ipynb | PhilipeRLeal/time_space_reductions | 8d826d0541846fac4e869db88c0b55794b32752f | [
"MIT"
] | null | null | null | 37.105948 | 352 | 0.441717 | [
[
[
"import numpy as np\nimport pandas as pd\nimport xarray as xr\nimport geopandas as gpd\nfrom shapely.geometry import Point",
"_____no_output_____"
],
[
"import sys\nimport os\nsys.path.insert(0, os.path.dirname(os.getcwd()))",
"_____no_output_____"
],
[
"from time_space_reductions.match_ups_over_polygons import get_zonal_match_up",
"_____no_output_____"
],
[
"\ndef make_fake_data(N=200):\n\n # creating example GeoDataframe for match-ups in EPSG 4326\n\n xx = np.random.randint(low=-60, high=-33, size=N)*1.105\n\n yy = np.random.randint(low=-4, high=20, size=N)*1.105\n\n df = pd.DataFrame({'lon':xx, 'lat':yy})\n\n\n df['geometry'] = df.apply(lambda x: Point(x['lon'], x['lat']), axis=1)\n\n\n gdf = gpd.GeoDataFrame(df, geometry='geometry', crs={'init':'epsg:4326'})\n\n gdf['Datetime'] = pd.date_range('2010-05-19', '2010-06-24', periods=gdf.shape[0])\n\n gdf.crs = {'init' :'epsg:4326'}\n\n\n return gdf",
"_____no_output_____"
],
[
"def get_netcdf_example():\n import glob\n cpath = r'C:\\Users\\Philipe Leal\\Dropbox\\Profissao\\Python\\OSGEO\\Matrizes\\NetCDF\\Time_Space_Concatenations\\time_space_reductions\\tests\\data'\n path_file = glob.glob(cpath + '/*.nc' )\n\n\n return xr.open_mfdataset(path_file[0])",
"_____no_output_____"
]
],
[
[
"from time_space_reductions.netcdf_gdf_setter import Base_class_space_time_netcdf_gdf\n\nclass Space_Time_Agg_over_polygons(Base_class_space_time_netcdf_gdf):\n \n def __init__(self, gdf, xarray_dataset=None, \n netcdf_temporal_coord_name='time',\n geo_series_temporal_attribute_name = 'Datetime',\n longitude_dimension='lon',\n latitude_dimension='lat',\n\t\t\t\t):\n \n \n '''\n Class description:\n ------------------\n \n This class is a base class for ensuring that the given netcdf is in conformity with the algorithm.\n \n Ex: the Netcdf has to be sorted in ascending order for all dimensions (ex: time ,longitude, latitude). \n Otherwise, the returned algorithm would return Nan values for all slices\n \n Also, it is mandatory for the user to define the longitude and latitude dimension Names (ex: 'lon', 'lat'),\n since there is no stadardization for defining these properties in the Netcdf files worldwide.\n \n \n \n Attributes:\n \n gdf (geodataframe):\n -----------------------\n The geodataframe object containing geometries to be analyzed.\n \n xarray_dataset (None): \n -----------------------\n \n the Xarry Netcdf object to be analyzed\n \n \n netcdf_temporal_coord_name (str = 'time'): \n -----------------------------------\n \n the name of the time dimension in the netcdf file\n \n \n geo_series_temporal_attribute_name(str = 'Datetime'):\n -----------------------------------\n \n the name of the time dimension in the geoseries file \n \n \n longitude_dimension (str = 'lon'): \n ----------------------------------\n \n the name of the longitude/horizontal dimension in the netcdf file\n \n \n latitude_dimension (str = 'lat'): \n ----------------------------------\n the name of the latitude/vertical dimension in the netcdf file\n \n\n \n '''\n \n Base_class_space_time_netcdf_gdf.__init__(self, xarray_dataset=xarray_dataset, \n netcdf_temporal_coord_name=netcdf_temporal_coord_name,\n geo_series_temporal_attribute_name = geo_series_temporal_attribute_name,\n longitude_dimension=longitude_dimension,\n latitude_dimension=latitude_dimension,\n )\n \n \n self.__netcdf_ds = xarray_dataset\n \n self.__gdf = gdf\n self.__geo_series_temporal_attribute_name = geo_series_temporal_attribute_name\n \n self.__netcdf_ds = self.netcdf_ds.sortby([self._temporal_coords, \n longitude_dimension,\n latitude_dimension])\n \n self.netcdf_ds = self._slice_bounding_box()\n \n \n @ property\n \n def netcdf_ds(self):\n \n return self.__netcdf_ds\n \n @ netcdf_ds.setter\n \n def netcdf_ds(self, new_netcdf_ds):\n '''\n This property-setter alters the former netcdf_ds for the new gdf provided\n \n \n '''\n \n self.__netcdf_ds = new_netcdf_ds\n \n \n \n @ property\n \n def gdf(self):\n \n return self.__gdf\n \n @ gdf.setter\n \n def gdf(self, new_gdf):\n '''\n This property-setter alters the former GDF for the new gdf provided\n \n \n '''\n \n self.__gdf = new_gdf\n \n \n def _slice_bounding_box(self):\n \n xmin, ymin, xmax, ymax = self.gdf.geometry.total_bounds\n \n \n dx = float(self.coord_resolution(self.spatial_coords['x']))\n \n dy = float(self.coord_resolution(self.spatial_coords['y']))\n \n xmin -= dx # to ensure full pixel slicing\n \n xmax += dx # to ensure full pixel slicing \n \n ymin -= dy # to ensure full pixel slicing \n \n ymax += dy # to ensure full pixel slicing \n \n \n result = self.netcdf_ds.sel({self.spatial_coords['x']:slice(xmin, xmax),\n self.spatial_coords['y']:slice(ymin, ymax)})\n \n return result\n \n \n def _slice_time_interval(self, time_init, final_time):\n \n \n result = self.netcdf_ds.sel({self._temporal_coords:slice(time_init, final_time)})\n \n\n return result\n \n \n \n \n def _make_time_space_aggregations(self, \n geoDataFrame, \n date_offset,\n netcdf_varnames, \n agg_functions):\n \n Tmin = geoDataFrame[self.__geo_series_temporal_attribute_name].min()\n \n Tmax = geoDataFrame[self.__geo_series_temporal_attribute_name].max()\n \n time_init = Tmin - date_offset\n \n final_time = Tmax + date_offset\n \n netcdf_sliced = self._slice_time_interval(time_init, final_time)\n \n netcdf_sliced_as_gpd_geodataframe = self.netcdf_to_gdf(netcdf_sliced)\n \n if not netcdf_sliced_as_gpd_geodataframe.empty:\n \n sjoined = gpd.sjoin(geoDataFrame, netcdf_sliced_as_gpd_geodataframe, how=\"left\", op='contains')\n \n sjoined_agg = sjoined[netcdf_varnames].agg(agg_functions)\n \n \n else:\n sjoined = geoDataFrame\n \n for key in netcdf_varnames:\n sjoined[key] = np.nan\n \n \n sjoined_agg = sjoined_agg.T\n \n sjoined_agg['period_sliced'] = time_init.strftime(\"%Y/%m/%d %H:%M:%S\") + ' <-> ' + final_time.strftime(\"%Y/%m/%d %H:%M:%S\")\n \n sjoined_agg.index.name = 'Variables'\n \n print(sjoined_agg)\n \n #sjoined_agg.index = geodataframe.index ?\n \n return sjoined_agg\n \n def _evaluate_space_time_agg(self, \n netcdf_varnames=['adg_443_qaa'], \n dict_of_windows=dict(time_window='1D'),\n agg_functions=['nanmean','nansum','nanstd'],\n verbose=True):\n \n \n date_offset = pd.tseries.frequencies.to_offset(dict_of_windows['time_window'])\n \n \n self.gdf2 = self.gdf.groupby(self.__geo_series_temporal_attribute_name).apply(lambda group: \n \n self._make_time_space_aggregations(group,\n date_offset=date_offset,\n netcdf_varnames=netcdf_varnames,\n agg_functions=agg_functions)\n \n )\n \n if self.gdf.index.name == None:\n\n self.gdf.index.name = 'index'\n\n idx_name = 'index'\n\n else:\n idx_name = self.gdf.index.name\n\n T = self.gdf2\n T[idx_name] = list(self.gdf.index) * (len(self.gdf2) // len(self.gdf))\n T = T.set_index('index', append=True, inplace=False).swaplevel(2, 0)\n\n self.gdf2 = self.gdf.merge(T, on=idx_name)\n\n \n \ndef _base(gdf,\n netcdf,\n netcdf_varnames =['adg_443_qaa'],\n netcdf_temporal_coord_name='time',\n geo_series_temporal_attribute_name = 'Datetime',\n longitude_dimension='lon',\n latitude_dimension='lat',\n dict_of_windows=dict(time_window='M'),\n agg_functions=['mean', 'max', 'min', 'std'],\n verbose=True):\n \n \n \n \n Match_Upper = Space_Time_Agg_over_polygons( gdf=gdf, \n xarray_dataset=netcdf, \n netcdf_temporal_coord_name=netcdf_temporal_coord_name,\n geo_series_temporal_attribute_name = geo_series_temporal_attribute_name,\n longitude_dimension=longitude_dimension,\n latitude_dimension=latitude_dimension)\n \n \n Match_Upper._evaluate_space_time_agg(netcdf_varnames=netcdf_varnames, \n dict_of_windows=dict_of_windows,\n agg_functions=agg_functions,\n verbose=verbose)\n \n \n \n \n return Match_Upper.gdf2\n \n \n\ndef get_zonal_match_up(netcdf, \n\t\t\t\t\t gdf, \n netcdf_varnames =['adg_443_qaa'],\n dict_of_windows=dict(time_window='5D'),\n agg_functions=['mean', 'max', 'min', 'std'],\n netcdf_temporal_coord_name='time',\n geo_series_temporal_attribute_name = 'Datetime',\n longitude_dimension='lon',\n latitude_dimension='lat',\n verbose=True):\n \n \"\"\"\n This function does Match - Up operations from centroids of Geoseries or GeoDataFrames over Netcdfs.\n \n\tAttributes:\n\t\n\t\tnetcdf (xarray Dataset/Dataarray):\n\t\t--------------------------------------------------------------------------\n\t\t\n\t\t\n\t\tgdf (geopandas GeoDataFrame):\n\t\t--------------------------------------------------------------------------\n\t\t\n\t\t\n\t\tnetcdf_varnames (list): a list containing the netcdf variable names to apply the aggregation.\n\n\t\t\tExample: netcdf_varnames=['adg_443_qaa']\n\t\t--------------------------------------------------------------------------\n\t\t\n\t\t\n\t\tdict_of_windows(dictionary)\n\t\t\n\t\t\n\t\t\tExample: dict_of_windows=dict(time_window='5D') # for 5 day window integration\n\t\t\t\n\t\t\t\tOther time integration options, follow pandas pattern (e.g.: 'Q', '3Y',...etc.)\n\t\t\t\t\t\t\t \n\t\t--------------------------------------------------------------------------\n\t\t\n\t\t\n\t\tagg_functions(list):\n\t\t\n\t\t\tExample: agg_functions = ['mean', 'max', 'min', 'std']\n\t\t\t\n\t\t--------------------------------------------------------------------------\n\t\t\n\t\t\n\t\tverbose (bool): it sets the function to verbose (or not). \n\t\t\n\t\t\tExample verbose=True\n\t\t--------------------------------------------------------------------------\n\t\t\n\t\t\n\t\t\n\tReturns:\n\t\t(geopandas GeoDataFrame)\n\t\t\n\t\n \"\"\"\n \n if isinstance(gdf.index, pd.MultiIndex):\n \n gdf = gdf.reset_index()\n return _base(gdf=gdf.copy(), \n netcdf=netcdf.copy(), \n netcdf_varnames=netcdf_varnames,\n dict_of_windows=dict_of_windows,\n agg_functions=agg_functions,\n verbose=verbose,\n netcdf_temporal_coord_name=netcdf_temporal_coord_name,\n geo_series_temporal_attribute_name = geo_series_temporal_attribute_name,\n longitude_dimension=longitude_dimension,\n latitude_dimension=latitude_dimension,\n )\n ",
"_____no_output_____"
],
[
"# Getting data",
"_____no_output_____"
]
],
[
[
"gdf = make_fake_data(3)\n\ngdf.geometry = gdf.geometry.buffer(1.15) # in degrees\n\nxnetcdf = get_netcdf_example()\n",
"C:\\Anaconda3\\envs\\Python_3.8\\lib\\site-packages\\pyproj\\crs\\crs.py:53: FutureWarning: '+init=<authority>:<code>' syntax is deprecated. '<authority>:<code>' is the preferred initialization method. When making the change, be mindful of axis order changes: https://pyproj4.github.io/pyproj/stable/gotchas.html#axis-order-changes-in-proj-6\n return _prepare_from_string(\" \".join(pjargs))\n<ipython-input-6-dece4842b27b>:3: UserWarning: Geometry is in a geographic CRS. Results from 'buffer' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.\n\n gdf.geometry = gdf.geometry.buffer(1.15) # in degrees\n"
]
],
[
[
"# Using the algorithm",
"_____no_output_____"
]
],
[
[
"xnetcdf['new_data'] = xnetcdf['adg_443_qaa'] * 5 - 15\n\nMatch_Upper = get_zonal_match_up(gdf=gdf, \n netcdf=xnetcdf,\n netcdf_varnames =['adg_443_qaa', 'new_data'],\n dict_of_windows=dict(time_window='1M'),\n agg_functions=['mean', 'max', 'min', 'std']\n\n )\n\nMatch_Upper",
" mean max min std period_sliced\nVariables \nadg_443_qaa NaN NaN NaN NaN 2010/04/30 00:00:00 <-> 2010/05/31 00:00:00\nnew_data NaN NaN NaN NaN 2010/04/30 00:00:00 <-> 2010/05/31 00:00:00\n mean max min std period_sliced\nVariables \nadg_443_qaa NaN NaN NaN NaN 2010/05/31 00:00:00 <-> 2010/06/30 00:00:00\nnew_data NaN NaN NaN NaN 2010/05/31 00:00:00 <-> 2010/06/30 00:00:00\n"
],
[
"Match_Upper",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0b5f1f2f9c287062c7d3a7c88cb92a2311d224c | 127,199 | ipynb | Jupyter Notebook | nlp/v1_scripts/supervised_UCI_adam256_save_embedding.ipynb | solislemuslab/dna-nn-theory | 9e996d5f453e1d620dadca0c276cb4a68e2b68e5 | [
"MIT"
] | 1 | 2021-06-02T22:27:46.000Z | 2021-06-02T22:27:46.000Z | nlp/v1_scripts/supervised_UCI_adam256_save_embedding.ipynb | solislemuslab/dna-nn-theory | 9e996d5f453e1d620dadca0c276cb4a68e2b68e5 | [
"MIT"
] | null | null | null | nlp/v1_scripts/supervised_UCI_adam256_save_embedding.ipynb | solislemuslab/dna-nn-theory | 9e996d5f453e1d620dadca0c276cb4a68e2b68e5 | [
"MIT"
] | 1 | 2020-07-08T19:53:30.000Z | 2020-07-08T19:53:30.000Z | 111.774165 | 57,302 | 0.681892 | [
[
[
"<a href=\"https://colab.research.google.com/github/csy99/dna-nn-theory/blob/master/supervised_UCI_adam256_save_embedding.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib\nimport matplotlib.pyplot as plt\nfrom itertools import product\nimport re\nimport time\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.manifold import TSNE\nimport tensorflow as tf\nfrom tensorflow import keras",
"_____no_output_____"
]
],
[
[
"# Read Data",
"_____no_output_____"
]
],
[
[
"!pip install PyDrive\nfrom google.colab import drive\ndrive.mount('/content/gdrive')",
"Requirement already satisfied: PyDrive in /usr/local/lib/python3.6/dist-packages (1.3.1)\nRequirement already satisfied: oauth2client>=4.0.0 in /usr/local/lib/python3.6/dist-packages (from PyDrive) (4.1.3)\nRequirement already satisfied: google-api-python-client>=1.2 in /usr/local/lib/python3.6/dist-packages (from PyDrive) (1.7.12)\nRequirement already satisfied: PyYAML>=3.0 in /usr/local/lib/python3.6/dist-packages (from PyDrive) (3.13)\nRequirement already satisfied: httplib2>=0.9.1 in /usr/local/lib/python3.6/dist-packages (from oauth2client>=4.0.0->PyDrive) (0.17.4)\nRequirement already satisfied: pyasn1>=0.1.7 in /usr/local/lib/python3.6/dist-packages (from oauth2client>=4.0.0->PyDrive) (0.4.8)\nRequirement already satisfied: rsa>=3.1.4 in /usr/local/lib/python3.6/dist-packages (from oauth2client>=4.0.0->PyDrive) (4.6)\nRequirement already satisfied: six>=1.6.1 in /usr/local/lib/python3.6/dist-packages (from oauth2client>=4.0.0->PyDrive) (1.15.0)\nRequirement already satisfied: pyasn1-modules>=0.0.5 in /usr/local/lib/python3.6/dist-packages (from oauth2client>=4.0.0->PyDrive) (0.2.8)\nRequirement already satisfied: google-auth-httplib2>=0.0.3 in /usr/local/lib/python3.6/dist-packages (from google-api-python-client>=1.2->PyDrive) (0.0.4)\nRequirement already satisfied: uritemplate<4dev,>=3.0.0 in /usr/local/lib/python3.6/dist-packages (from google-api-python-client>=1.2->PyDrive) (3.0.1)\nRequirement already satisfied: google-auth>=1.4.1 in /usr/local/lib/python3.6/dist-packages (from google-api-python-client>=1.2->PyDrive) (1.17.2)\nRequirement already satisfied: setuptools>=40.3.0 in /usr/local/lib/python3.6/dist-packages (from google-auth>=1.4.1->google-api-python-client>=1.2->PyDrive) (50.3.2)\nRequirement already satisfied: cachetools<5.0,>=2.0.0 in /usr/local/lib/python3.6/dist-packages (from google-auth>=1.4.1->google-api-python-client>=1.2->PyDrive) (4.1.1)\nMounted at /content/gdrive\n"
],
[
"def convert_label(row):\n if row[\"Classes\"] == 'EI':\n return 0\n if row[\"Classes\"] == 'IE':\n return 1\n if row[\"Classes\"] == 'N':\n return 2",
"_____no_output_____"
],
[
"data_path = '/content/gdrive/My Drive/Colab Notebooks/UCI/'\nsplice_df = pd.read_csv(data_path + 'splice.data', header=None)\nsplice_df.columns = ['Classes', 'Name', 'Seq']\nsplice_df[\"Seq\"] = splice_df[\"Seq\"].str.replace(' ', '').str.replace('N', 'A').str.replace('D', 'T').str.replace('S', 'C').str.replace('R', 'G')\nsplice_df[\"Label\"] = splice_df.apply(lambda row: convert_label(row), axis=1)\nprint('The shape of the datasize is', splice_df.shape)\nsplice_df.head()",
"The shape of the datasize is (3190, 4)\n"
],
[
"seq_num = 0\nfor seq in splice_df[\"Seq\"]:\n char_num = 0\n for char in seq:\n if char != 'A' and char != 'C' and char != 'T' and char != 'G':\n print(\"seq\", seq_num, 'char', char_num, 'is', char)\n char_num += 1\n seq_num += 1",
"_____no_output_____"
],
[
"# check if the length of the sequence is the same \nseq_len = len(splice_df.Seq[0])\nprint(\"The length of the sequence is\", seq_len)\nfor seq in splice_df.Seq[:200]:\n assert len(seq) == seq_len",
"The length of the sequence is 60\n"
],
[
"xtrain_full, xtest, ytrain_full, ytest = train_test_split(splice_df, splice_df.Label, test_size=0.2, random_state=100, stratify=splice_df.Label)\nxtrain, xval, ytrain, yval = train_test_split(xtrain_full, ytrain_full, test_size=0.2, random_state=100, stratify=ytrain_full)\nprint(\"shape of training, validation, test set\\n\", xtrain.shape, xval.shape, xtest.shape, ytrain.shape, yval.shape, ytest.shape)",
"shape of training, validation, test set\n (2041, 4) (511, 4) (638, 4) (2041,) (511,) (638,)\n"
],
[
"word_size = 1\nvocab = [''.join(p) for p in product('ACGT', repeat=word_size)]\nword_to_idx = {word: i for i, word in enumerate(vocab)}\nvocab_size = len(word_to_idx)\nprint('vocab_size:', vocab_size)\ncreate1gram = keras.layers.experimental.preprocessing.TextVectorization(\n standardize=lambda x: tf.strings.regex_replace(x, '(.)', '\\\\1 '), ngrams=1\n)\ncreate1gram.adapt(vocab)",
"vocab_size: 4\n"
],
[
"def ds_preprocess(x, y):\n x_index = tf.subtract(create1gram(x), 2)\n return x_index, y",
"_____no_output_____"
],
[
"# not sure the correct way to get mapping from word to its index\ncreate1gram('A C G T') - 2",
"_____no_output_____"
],
[
"BATCH_SIZE = 256\nxtrain_ds = tf.data.Dataset.from_tensor_slices((xtrain['Seq'], ytrain)).map(ds_preprocess).batch(BATCH_SIZE)\nxval_ds = tf.data.Dataset.from_tensor_slices((xval['Seq'], yval)).map(ds_preprocess).batch(BATCH_SIZE)\nxtest_ds = tf.data.Dataset.from_tensor_slices((xtest['Seq'], ytest)).map(ds_preprocess).batch(BATCH_SIZE)",
"_____no_output_____"
],
[
"latent_size = 30\n\nmodel = keras.Sequential([\n keras.Input(shape=(seq_len,)),\n keras.layers.Embedding(seq_len, latent_size),\n keras.layers.LSTM(latent_size, return_sequences=False),\n keras.layers.Dense(128, activation=\"relu\", input_shape=[latent_size]),\n keras.layers.Dropout(0.2),\n keras.layers.Dense(64, activation=\"relu\"), \n keras.layers.Dropout(0.2),\n keras.layers.Dense(32, activation=\"relu\"), \n keras.layers.Dropout(0.2), \n keras.layers.Dense(16, activation=\"relu\"), \n keras.layers.Dropout(0.2), \n keras.layers.Dense(3, activation=\"softmax\") \n])\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, 60, 30) 1800 \n_________________________________________________________________\nlstm (LSTM) (None, 30) 7320 \n_________________________________________________________________\ndense (Dense) (None, 128) 3968 \n_________________________________________________________________\ndropout (Dropout) (None, 128) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 64) 8256 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 64) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 32) 2080 \n_________________________________________________________________\ndropout_2 (Dropout) (None, 32) 0 \n_________________________________________________________________\ndense_3 (Dense) (None, 16) 528 \n_________________________________________________________________\ndropout_3 (Dropout) (None, 16) 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 3) 51 \n=================================================================\nTotal params: 24,003\nTrainable params: 24,003\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"es_cb = keras.callbacks.EarlyStopping(patience=100, restore_best_weights=True)\nmodel.compile(keras.optimizers.Adam(), loss=keras.losses.SparseCategoricalCrossentropy(), metrics=['accuracy'])\nhist = model.fit(xtrain_ds, validation_data=xval_ds, epochs=4000, callbacks=[es_cb])",
"Epoch 1/4000\n8/8 [==============================] - 1s 104ms/step - loss: 1.0801 - accuracy: 0.5076 - val_loss: 1.0591 - val_accuracy: 0.5186\nEpoch 2/4000\n8/8 [==============================] - 0s 56ms/step - loss: 1.0436 - accuracy: 0.5149 - val_loss: 1.0287 - val_accuracy: 0.5186\nEpoch 3/4000\n8/8 [==============================] - 0s 57ms/step - loss: 1.0314 - accuracy: 0.5189 - val_loss: 1.0241 - val_accuracy: 0.5186\nEpoch 4/4000\n8/8 [==============================] - 0s 57ms/step - loss: 1.0342 - accuracy: 0.5189 - val_loss: 1.0255 - val_accuracy: 0.5186\nEpoch 5/4000\n8/8 [==============================] - 0s 59ms/step - loss: 1.0278 - accuracy: 0.5189 - val_loss: 1.0192 - val_accuracy: 0.5186\nEpoch 6/4000\n8/8 [==============================] - 0s 62ms/step - loss: 1.0293 - accuracy: 0.5189 - val_loss: 1.0159 - val_accuracy: 0.5186\nEpoch 7/4000\n8/8 [==============================] - 0s 56ms/step - loss: 1.0225 - accuracy: 0.5189 - val_loss: 1.0082 - val_accuracy: 0.5186\nEpoch 8/4000\n8/8 [==============================] - 0s 59ms/step - loss: 1.0176 - accuracy: 0.5189 - val_loss: 1.0036 - val_accuracy: 0.5186\nEpoch 9/4000\n8/8 [==============================] - 0s 58ms/step - loss: 1.0049 - accuracy: 0.5189 - val_loss: 0.9980 - val_accuracy: 0.5186\nEpoch 10/4000\n8/8 [==============================] - 0s 58ms/step - loss: 1.0045 - accuracy: 0.5203 - val_loss: 0.9941 - val_accuracy: 0.5186\nEpoch 11/4000\n8/8 [==============================] - 0s 58ms/step - loss: 1.0019 - accuracy: 0.5198 - val_loss: 0.9896 - val_accuracy: 0.5186\nEpoch 12/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.9866 - accuracy: 0.5179 - val_loss: 0.9775 - val_accuracy: 0.5186\nEpoch 13/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.9786 - accuracy: 0.5252 - val_loss: 0.9685 - val_accuracy: 0.5401\nEpoch 14/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.9817 - accuracy: 0.5262 - val_loss: 0.9680 - val_accuracy: 0.5616\nEpoch 15/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.9755 - accuracy: 0.5233 - val_loss: 0.9622 - val_accuracy: 0.5577\nEpoch 16/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.9598 - accuracy: 0.5203 - val_loss: 0.9500 - val_accuracy: 0.5362\nEpoch 17/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.9337 - accuracy: 0.5145 - val_loss: 0.9114 - val_accuracy: 0.5538\nEpoch 18/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.9081 - accuracy: 0.5233 - val_loss: 0.8865 - val_accuracy: 0.5538\nEpoch 19/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.8834 - accuracy: 0.5331 - val_loss: 0.8764 - val_accuracy: 0.5440\nEpoch 20/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.8520 - accuracy: 0.5576 - val_loss: 0.8283 - val_accuracy: 0.5988\nEpoch 21/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.8274 - accuracy: 0.5884 - val_loss: 0.7993 - val_accuracy: 0.6204\nEpoch 22/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.8036 - accuracy: 0.6017 - val_loss: 0.8033 - val_accuracy: 0.6164\nEpoch 23/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.7831 - accuracy: 0.6198 - val_loss: 0.7872 - val_accuracy: 0.6145\nEpoch 24/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.7683 - accuracy: 0.6389 - val_loss: 0.7775 - val_accuracy: 0.6399\nEpoch 25/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.7748 - accuracy: 0.6232 - val_loss: 0.7710 - val_accuracy: 0.6321\nEpoch 26/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.7603 - accuracy: 0.6267 - val_loss: 0.7695 - val_accuracy: 0.6301\nEpoch 27/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.7626 - accuracy: 0.6335 - val_loss: 0.8252 - val_accuracy: 0.6047\nEpoch 28/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.7566 - accuracy: 0.6335 - val_loss: 0.7667 - val_accuracy: 0.6282\nEpoch 29/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.7554 - accuracy: 0.6394 - val_loss: 0.7583 - val_accuracy: 0.6399\nEpoch 30/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.7276 - accuracy: 0.6472 - val_loss: 0.7756 - val_accuracy: 0.6438\nEpoch 31/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.7198 - accuracy: 0.6575 - val_loss: 0.7875 - val_accuracy: 0.6517\nEpoch 32/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.7226 - accuracy: 0.6614 - val_loss: 0.7707 - val_accuracy: 0.6575\nEpoch 33/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.7203 - accuracy: 0.6477 - val_loss: 0.7579 - val_accuracy: 0.6575\nEpoch 34/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.7146 - accuracy: 0.6531 - val_loss: 0.7526 - val_accuracy: 0.6732\nEpoch 35/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.7080 - accuracy: 0.6610 - val_loss: 0.7510 - val_accuracy: 0.6673\nEpoch 36/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.7125 - accuracy: 0.6634 - val_loss: 0.7731 - val_accuracy: 0.6634\nEpoch 37/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.7126 - accuracy: 0.6707 - val_loss: 0.8103 - val_accuracy: 0.6595\nEpoch 38/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.7032 - accuracy: 0.6619 - val_loss: 0.7598 - val_accuracy: 0.6712\nEpoch 39/4000\n8/8 [==============================] - 1s 64ms/step - loss: 0.6977 - accuracy: 0.6801 - val_loss: 0.7432 - val_accuracy: 0.6712\nEpoch 40/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.6931 - accuracy: 0.6678 - val_loss: 0.7439 - val_accuracy: 0.6751\nEpoch 41/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.6736 - accuracy: 0.6801 - val_loss: 0.7535 - val_accuracy: 0.6908\nEpoch 42/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.6813 - accuracy: 0.6683 - val_loss: 0.7722 - val_accuracy: 0.6791\nEpoch 43/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.6851 - accuracy: 0.6889 - val_loss: 0.7619 - val_accuracy: 0.6830\nEpoch 44/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.6759 - accuracy: 0.6830 - val_loss: 0.7901 - val_accuracy: 0.6791\nEpoch 45/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.6847 - accuracy: 0.6786 - val_loss: 0.7589 - val_accuracy: 0.6830\nEpoch 46/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.6771 - accuracy: 0.6864 - val_loss: 0.7195 - val_accuracy: 0.7065\nEpoch 47/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.6635 - accuracy: 0.6908 - val_loss: 0.7131 - val_accuracy: 0.7006\nEpoch 48/4000\n8/8 [==============================] - 1s 64ms/step - loss: 0.6491 - accuracy: 0.7021 - val_loss: 0.7161 - val_accuracy: 0.7123\nEpoch 49/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.6402 - accuracy: 0.7129 - val_loss: 0.7150 - val_accuracy: 0.7025\nEpoch 50/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.6365 - accuracy: 0.7139 - val_loss: 0.7233 - val_accuracy: 0.7162\nEpoch 51/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.6482 - accuracy: 0.7021 - val_loss: 0.7328 - val_accuracy: 0.7123\nEpoch 52/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.6349 - accuracy: 0.7124 - val_loss: 0.7357 - val_accuracy: 0.7065\nEpoch 53/4000\n8/8 [==============================] - 1s 65ms/step - loss: 0.6407 - accuracy: 0.7065 - val_loss: 0.7424 - val_accuracy: 0.6849\nEpoch 54/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.6576 - accuracy: 0.6967 - val_loss: 0.6853 - val_accuracy: 0.7104\nEpoch 55/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.6280 - accuracy: 0.7222 - val_loss: 0.6846 - val_accuracy: 0.7084\nEpoch 56/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.6039 - accuracy: 0.7295 - val_loss: 0.6994 - val_accuracy: 0.6928\nEpoch 57/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.6114 - accuracy: 0.7349 - val_loss: 0.6940 - val_accuracy: 0.7065\nEpoch 58/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.6106 - accuracy: 0.7227 - val_loss: 0.6792 - val_accuracy: 0.7202\nEpoch 59/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.6129 - accuracy: 0.7286 - val_loss: 0.6837 - val_accuracy: 0.7319\nEpoch 60/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.6071 - accuracy: 0.7310 - val_loss: 0.7077 - val_accuracy: 0.7182\nEpoch 61/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.6147 - accuracy: 0.7266 - val_loss: 0.6761 - val_accuracy: 0.7143\nEpoch 62/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.6139 - accuracy: 0.7320 - val_loss: 0.6701 - val_accuracy: 0.7025\nEpoch 63/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.5978 - accuracy: 0.7344 - val_loss: 0.6680 - val_accuracy: 0.7241\nEpoch 64/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5972 - accuracy: 0.7393 - val_loss: 0.6627 - val_accuracy: 0.7162\nEpoch 65/4000\n8/8 [==============================] - 0s 62ms/step - loss: 0.5933 - accuracy: 0.7438 - val_loss: 0.6644 - val_accuracy: 0.7143\nEpoch 66/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5944 - accuracy: 0.7364 - val_loss: 0.6676 - val_accuracy: 0.7143\nEpoch 67/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5926 - accuracy: 0.7487 - val_loss: 0.6579 - val_accuracy: 0.7182\nEpoch 68/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.5911 - accuracy: 0.7408 - val_loss: 0.6588 - val_accuracy: 0.7241\nEpoch 69/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.5769 - accuracy: 0.7408 - val_loss: 0.6619 - val_accuracy: 0.7065\nEpoch 70/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.5799 - accuracy: 0.7482 - val_loss: 0.6543 - val_accuracy: 0.7143\nEpoch 71/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5573 - accuracy: 0.7599 - val_loss: 0.6553 - val_accuracy: 0.7182\nEpoch 72/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5707 - accuracy: 0.7452 - val_loss: 0.6463 - val_accuracy: 0.7221\nEpoch 73/4000\n8/8 [==============================] - 0s 62ms/step - loss: 0.5689 - accuracy: 0.7545 - val_loss: 0.6439 - val_accuracy: 0.7280\nEpoch 74/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5678 - accuracy: 0.7472 - val_loss: 0.6346 - val_accuracy: 0.7202\nEpoch 75/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5662 - accuracy: 0.7531 - val_loss: 0.6414 - val_accuracy: 0.7104\nEpoch 76/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5690 - accuracy: 0.7540 - val_loss: 0.6388 - val_accuracy: 0.7202\nEpoch 77/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5512 - accuracy: 0.7634 - val_loss: 0.6398 - val_accuracy: 0.7202\nEpoch 78/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.5472 - accuracy: 0.7673 - val_loss: 0.6409 - val_accuracy: 0.7241\nEpoch 79/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.5367 - accuracy: 0.7663 - val_loss: 0.6448 - val_accuracy: 0.7260\nEpoch 80/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5442 - accuracy: 0.7692 - val_loss: 0.6362 - val_accuracy: 0.7202\nEpoch 81/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5331 - accuracy: 0.7638 - val_loss: 0.6366 - val_accuracy: 0.7319\nEpoch 82/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5403 - accuracy: 0.7697 - val_loss: 0.6301 - val_accuracy: 0.7202\nEpoch 83/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.5472 - accuracy: 0.7653 - val_loss: 0.6298 - val_accuracy: 0.7378\nEpoch 84/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5445 - accuracy: 0.7722 - val_loss: 0.6264 - val_accuracy: 0.7241\nEpoch 85/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5322 - accuracy: 0.7741 - val_loss: 0.6292 - val_accuracy: 0.7280\nEpoch 86/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5307 - accuracy: 0.7687 - val_loss: 0.6184 - val_accuracy: 0.7319\nEpoch 87/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5332 - accuracy: 0.7756 - val_loss: 0.6112 - val_accuracy: 0.7476\nEpoch 88/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.5149 - accuracy: 0.7829 - val_loss: 0.6268 - val_accuracy: 0.7378\nEpoch 89/4000\n8/8 [==============================] - 0s 54ms/step - loss: 0.5236 - accuracy: 0.7673 - val_loss: 0.6201 - val_accuracy: 0.7417\nEpoch 90/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5178 - accuracy: 0.7834 - val_loss: 0.6437 - val_accuracy: 0.7260\nEpoch 91/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5337 - accuracy: 0.7727 - val_loss: 0.6429 - val_accuracy: 0.7319\nEpoch 92/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.5289 - accuracy: 0.7727 - val_loss: 0.6527 - val_accuracy: 0.7143\nEpoch 93/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.5305 - accuracy: 0.7795 - val_loss: 0.6703 - val_accuracy: 0.7182\nEpoch 94/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5230 - accuracy: 0.7697 - val_loss: 0.6576 - val_accuracy: 0.7221\nEpoch 95/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.5276 - accuracy: 0.7805 - val_loss: 0.6954 - val_accuracy: 0.7123\nEpoch 96/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5349 - accuracy: 0.7746 - val_loss: 0.6655 - val_accuracy: 0.7202\nEpoch 97/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5220 - accuracy: 0.7702 - val_loss: 0.6405 - val_accuracy: 0.7299\nEpoch 98/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5242 - accuracy: 0.7732 - val_loss: 0.6086 - val_accuracy: 0.7397\nEpoch 99/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5209 - accuracy: 0.7776 - val_loss: 0.6231 - val_accuracy: 0.7202\nEpoch 100/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.5140 - accuracy: 0.7785 - val_loss: 0.6183 - val_accuracy: 0.7280\nEpoch 101/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.5111 - accuracy: 0.7839 - val_loss: 0.6228 - val_accuracy: 0.7397\nEpoch 102/4000\n8/8 [==============================] - 1s 70ms/step - loss: 0.5083 - accuracy: 0.7751 - val_loss: 0.6187 - val_accuracy: 0.7339\nEpoch 103/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4964 - accuracy: 0.7776 - val_loss: 0.6432 - val_accuracy: 0.7417\nEpoch 104/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4906 - accuracy: 0.7810 - val_loss: 0.6560 - val_accuracy: 0.7436\nEpoch 105/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4974 - accuracy: 0.7893 - val_loss: 0.6561 - val_accuracy: 0.7378\nEpoch 106/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4868 - accuracy: 0.7820 - val_loss: 0.6399 - val_accuracy: 0.7436\nEpoch 107/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4946 - accuracy: 0.7825 - val_loss: 0.6616 - val_accuracy: 0.7260\nEpoch 108/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5153 - accuracy: 0.7810 - val_loss: 0.6380 - val_accuracy: 0.7358\nEpoch 109/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.4992 - accuracy: 0.7776 - val_loss: 0.6128 - val_accuracy: 0.7436\nEpoch 110/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4996 - accuracy: 0.7844 - val_loss: 0.6178 - val_accuracy: 0.7378\nEpoch 111/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.5091 - accuracy: 0.7805 - val_loss: 0.6189 - val_accuracy: 0.7319\nEpoch 112/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.4942 - accuracy: 0.7864 - val_loss: 0.6329 - val_accuracy: 0.7260\nEpoch 113/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.5122 - accuracy: 0.7825 - val_loss: 0.6289 - val_accuracy: 0.7378\nEpoch 114/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.5145 - accuracy: 0.7834 - val_loss: 0.6307 - val_accuracy: 0.7241\nEpoch 115/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4888 - accuracy: 0.7942 - val_loss: 0.6476 - val_accuracy: 0.7417\nEpoch 116/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.4728 - accuracy: 0.7981 - val_loss: 0.6244 - val_accuracy: 0.7358\nEpoch 117/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.4778 - accuracy: 0.7991 - val_loss: 0.6061 - val_accuracy: 0.7476\nEpoch 118/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4671 - accuracy: 0.7976 - val_loss: 0.6156 - val_accuracy: 0.7495\nEpoch 119/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4652 - accuracy: 0.7937 - val_loss: 0.6203 - val_accuracy: 0.7339\nEpoch 120/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4713 - accuracy: 0.8030 - val_loss: 0.6001 - val_accuracy: 0.7554\nEpoch 121/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4633 - accuracy: 0.8011 - val_loss: 0.6090 - val_accuracy: 0.7436\nEpoch 122/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4581 - accuracy: 0.7927 - val_loss: 0.6103 - val_accuracy: 0.7593\nEpoch 123/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4550 - accuracy: 0.8021 - val_loss: 0.5957 - val_accuracy: 0.7534\nEpoch 124/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4592 - accuracy: 0.8011 - val_loss: 0.6302 - val_accuracy: 0.7397\nEpoch 125/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4494 - accuracy: 0.8109 - val_loss: 0.5954 - val_accuracy: 0.7476\nEpoch 126/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4543 - accuracy: 0.8060 - val_loss: 0.6001 - val_accuracy: 0.7534\nEpoch 127/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4636 - accuracy: 0.8006 - val_loss: 0.6092 - val_accuracy: 0.7515\nEpoch 128/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4366 - accuracy: 0.8168 - val_loss: 0.6039 - val_accuracy: 0.7573\nEpoch 129/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4460 - accuracy: 0.8109 - val_loss: 0.6233 - val_accuracy: 0.7476\nEpoch 130/4000\n8/8 [==============================] - 0s 62ms/step - loss: 0.4480 - accuracy: 0.8123 - val_loss: 0.5993 - val_accuracy: 0.7397\nEpoch 131/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4369 - accuracy: 0.8182 - val_loss: 0.5912 - val_accuracy: 0.7515\nEpoch 132/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.4324 - accuracy: 0.8187 - val_loss: 0.5874 - val_accuracy: 0.7456\nEpoch 133/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4200 - accuracy: 0.8138 - val_loss: 0.5982 - val_accuracy: 0.7632\nEpoch 134/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4248 - accuracy: 0.8192 - val_loss: 0.6006 - val_accuracy: 0.7515\nEpoch 135/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4389 - accuracy: 0.8221 - val_loss: 0.6046 - val_accuracy: 0.7495\nEpoch 136/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4371 - accuracy: 0.8241 - val_loss: 0.6113 - val_accuracy: 0.7652\nEpoch 137/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4304 - accuracy: 0.8133 - val_loss: 0.6044 - val_accuracy: 0.7671\nEpoch 138/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.4395 - accuracy: 0.8217 - val_loss: 0.5941 - val_accuracy: 0.7613\nEpoch 139/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.4386 - accuracy: 0.8236 - val_loss: 0.6142 - val_accuracy: 0.7613\nEpoch 140/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4444 - accuracy: 0.8182 - val_loss: 0.5752 - val_accuracy: 0.7593\nEpoch 141/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4219 - accuracy: 0.8275 - val_loss: 0.5831 - val_accuracy: 0.7730\nEpoch 142/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4164 - accuracy: 0.8364 - val_loss: 0.5945 - val_accuracy: 0.7613\nEpoch 143/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4113 - accuracy: 0.8315 - val_loss: 0.6012 - val_accuracy: 0.7573\nEpoch 144/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4172 - accuracy: 0.8290 - val_loss: 0.5939 - val_accuracy: 0.7573\nEpoch 145/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4110 - accuracy: 0.8339 - val_loss: 0.5794 - val_accuracy: 0.7808\nEpoch 146/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.3982 - accuracy: 0.8339 - val_loss: 0.5773 - val_accuracy: 0.7730\nEpoch 147/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.4040 - accuracy: 0.8349 - val_loss: 0.5777 - val_accuracy: 0.7808\nEpoch 148/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3945 - accuracy: 0.8359 - val_loss: 0.5863 - val_accuracy: 0.7691\nEpoch 149/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3866 - accuracy: 0.8417 - val_loss: 0.5823 - val_accuracy: 0.7828\nEpoch 150/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4083 - accuracy: 0.8319 - val_loss: 0.5810 - val_accuracy: 0.7750\nEpoch 151/4000\n8/8 [==============================] - 0s 55ms/step - loss: 0.3974 - accuracy: 0.8339 - val_loss: 0.5974 - val_accuracy: 0.7847\nEpoch 152/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.4188 - accuracy: 0.8241 - val_loss: 0.5903 - val_accuracy: 0.7710\nEpoch 153/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4045 - accuracy: 0.8290 - val_loss: 0.5889 - val_accuracy: 0.7945\nEpoch 154/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3855 - accuracy: 0.8373 - val_loss: 0.5909 - val_accuracy: 0.7730\nEpoch 155/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3950 - accuracy: 0.8457 - val_loss: 0.5944 - val_accuracy: 0.7828\nEpoch 156/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.4112 - accuracy: 0.8319 - val_loss: 0.5726 - val_accuracy: 0.7593\nEpoch 157/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3928 - accuracy: 0.8408 - val_loss: 0.5757 - val_accuracy: 0.7808\nEpoch 158/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3844 - accuracy: 0.8408 - val_loss: 0.5897 - val_accuracy: 0.7710\nEpoch 159/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3758 - accuracy: 0.8476 - val_loss: 0.5878 - val_accuracy: 0.7730\nEpoch 160/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3772 - accuracy: 0.8476 - val_loss: 0.5662 - val_accuracy: 0.7847\nEpoch 161/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3936 - accuracy: 0.8403 - val_loss: 0.5676 - val_accuracy: 0.7808\nEpoch 162/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3828 - accuracy: 0.8422 - val_loss: 0.5873 - val_accuracy: 0.7750\nEpoch 163/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3734 - accuracy: 0.8481 - val_loss: 0.6071 - val_accuracy: 0.7789\nEpoch 164/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3841 - accuracy: 0.8427 - val_loss: 0.5835 - val_accuracy: 0.7828\nEpoch 165/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3649 - accuracy: 0.8550 - val_loss: 0.5697 - val_accuracy: 0.7926\nEpoch 166/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3716 - accuracy: 0.8476 - val_loss: 0.6263 - val_accuracy: 0.7769\nEpoch 167/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3820 - accuracy: 0.8525 - val_loss: 0.5926 - val_accuracy: 0.7769\nEpoch 168/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3535 - accuracy: 0.8574 - val_loss: 0.5816 - val_accuracy: 0.7886\nEpoch 169/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3582 - accuracy: 0.8594 - val_loss: 0.6086 - val_accuracy: 0.7710\nEpoch 170/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3627 - accuracy: 0.8520 - val_loss: 0.6300 - val_accuracy: 0.7730\nEpoch 171/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.4103 - accuracy: 0.8383 - val_loss: 0.6116 - val_accuracy: 0.7691\nEpoch 172/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4058 - accuracy: 0.8417 - val_loss: 0.6302 - val_accuracy: 0.7710\nEpoch 173/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3965 - accuracy: 0.8378 - val_loss: 0.5679 - val_accuracy: 0.7808\nEpoch 174/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.4080 - accuracy: 0.8334 - val_loss: 0.5738 - val_accuracy: 0.7769\nEpoch 175/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.4404 - accuracy: 0.8251 - val_loss: 0.5564 - val_accuracy: 0.7828\nEpoch 176/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4195 - accuracy: 0.8339 - val_loss: 0.6405 - val_accuracy: 0.7573\nEpoch 177/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4389 - accuracy: 0.8266 - val_loss: 0.6156 - val_accuracy: 0.7613\nEpoch 178/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3956 - accuracy: 0.8373 - val_loss: 0.6001 - val_accuracy: 0.7867\nEpoch 179/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3664 - accuracy: 0.8496 - val_loss: 0.5711 - val_accuracy: 0.7984\nEpoch 180/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3676 - accuracy: 0.8604 - val_loss: 0.5861 - val_accuracy: 0.7828\nEpoch 181/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3453 - accuracy: 0.8643 - val_loss: 0.6006 - val_accuracy: 0.7926\nEpoch 182/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3406 - accuracy: 0.8682 - val_loss: 0.5801 - val_accuracy: 0.7926\nEpoch 183/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3436 - accuracy: 0.8711 - val_loss: 0.5703 - val_accuracy: 0.7945\nEpoch 184/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3175 - accuracy: 0.8775 - val_loss: 0.5932 - val_accuracy: 0.7965\nEpoch 185/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3655 - accuracy: 0.8653 - val_loss: 0.5738 - val_accuracy: 0.7945\nEpoch 186/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3818 - accuracy: 0.8462 - val_loss: 0.6063 - val_accuracy: 0.7926\nEpoch 187/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3695 - accuracy: 0.8535 - val_loss: 0.5682 - val_accuracy: 0.7886\nEpoch 188/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3903 - accuracy: 0.8447 - val_loss: 0.6898 - val_accuracy: 0.7691\nEpoch 189/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.4093 - accuracy: 0.8437 - val_loss: 0.6256 - val_accuracy: 0.7691\nEpoch 190/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3774 - accuracy: 0.8496 - val_loss: 0.5690 - val_accuracy: 0.7789\nEpoch 191/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3410 - accuracy: 0.8682 - val_loss: 0.5808 - val_accuracy: 0.7906\nEpoch 192/4000\n8/8 [==============================] - 0s 54ms/step - loss: 0.3263 - accuracy: 0.8736 - val_loss: 0.5781 - val_accuracy: 0.7945\nEpoch 193/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3089 - accuracy: 0.8780 - val_loss: 0.6007 - val_accuracy: 0.7926\nEpoch 194/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3328 - accuracy: 0.8707 - val_loss: 0.6415 - val_accuracy: 0.7808\nEpoch 195/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3290 - accuracy: 0.8736 - val_loss: 0.6723 - val_accuracy: 0.7847\nEpoch 196/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3485 - accuracy: 0.8658 - val_loss: 0.6504 - val_accuracy: 0.7750\nEpoch 197/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3310 - accuracy: 0.8687 - val_loss: 0.6300 - val_accuracy: 0.7867\nEpoch 198/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3256 - accuracy: 0.8760 - val_loss: 0.6210 - val_accuracy: 0.7906\nEpoch 199/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3226 - accuracy: 0.8790 - val_loss: 0.5685 - val_accuracy: 0.8023\nEpoch 200/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3223 - accuracy: 0.8775 - val_loss: 0.5887 - val_accuracy: 0.8004\nEpoch 201/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2995 - accuracy: 0.8888 - val_loss: 0.5669 - val_accuracy: 0.7926\nEpoch 202/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2943 - accuracy: 0.8854 - val_loss: 0.5959 - val_accuracy: 0.8063\nEpoch 203/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.2960 - accuracy: 0.8898 - val_loss: 0.5958 - val_accuracy: 0.7945\nEpoch 204/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.2992 - accuracy: 0.8863 - val_loss: 0.5911 - val_accuracy: 0.8023\nEpoch 205/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.3040 - accuracy: 0.8849 - val_loss: 0.5853 - val_accuracy: 0.8043\nEpoch 206/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3066 - accuracy: 0.8824 - val_loss: 0.6292 - val_accuracy: 0.7984\nEpoch 207/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3276 - accuracy: 0.8721 - val_loss: 0.5860 - val_accuracy: 0.7945\nEpoch 208/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3193 - accuracy: 0.8849 - val_loss: 0.6110 - val_accuracy: 0.7926\nEpoch 209/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3311 - accuracy: 0.8707 - val_loss: 0.5781 - val_accuracy: 0.8043\nEpoch 210/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.3247 - accuracy: 0.8751 - val_loss: 0.6857 - val_accuracy: 0.7886\nEpoch 211/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3297 - accuracy: 0.8785 - val_loss: 0.6486 - val_accuracy: 0.7808\nEpoch 212/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3488 - accuracy: 0.8697 - val_loss: 0.6569 - val_accuracy: 0.7730\nEpoch 213/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.3305 - accuracy: 0.8716 - val_loss: 0.5736 - val_accuracy: 0.7945\nEpoch 214/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.3038 - accuracy: 0.8873 - val_loss: 0.5831 - val_accuracy: 0.8043\nEpoch 215/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3038 - accuracy: 0.8854 - val_loss: 0.5827 - val_accuracy: 0.8043\nEpoch 216/4000\n8/8 [==============================] - 0s 53ms/step - loss: 0.2914 - accuracy: 0.8932 - val_loss: 0.6263 - val_accuracy: 0.7965\nEpoch 217/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2795 - accuracy: 0.8981 - val_loss: 0.6207 - val_accuracy: 0.8023\nEpoch 218/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2634 - accuracy: 0.9005 - val_loss: 0.6407 - val_accuracy: 0.8004\nEpoch 219/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2702 - accuracy: 0.9035 - val_loss: 0.6351 - val_accuracy: 0.8121\nEpoch 220/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2582 - accuracy: 0.9035 - val_loss: 0.5995 - val_accuracy: 0.8121\nEpoch 221/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.2645 - accuracy: 0.9040 - val_loss: 0.6145 - val_accuracy: 0.8102\nEpoch 222/4000\n8/8 [==============================] - 1s 74ms/step - loss: 0.2502 - accuracy: 0.9103 - val_loss: 0.6169 - val_accuracy: 0.8102\nEpoch 223/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2611 - accuracy: 0.9064 - val_loss: 0.6318 - val_accuracy: 0.8023\nEpoch 224/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2572 - accuracy: 0.9054 - val_loss: 0.6470 - val_accuracy: 0.8219\nEpoch 225/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2598 - accuracy: 0.9059 - val_loss: 0.5903 - val_accuracy: 0.8082\nEpoch 226/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2620 - accuracy: 0.9045 - val_loss: 0.6819 - val_accuracy: 0.8063\nEpoch 227/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2714 - accuracy: 0.9020 - val_loss: 0.6737 - val_accuracy: 0.8004\nEpoch 228/4000\n8/8 [==============================] - 0s 55ms/step - loss: 0.2778 - accuracy: 0.8951 - val_loss: 0.5933 - val_accuracy: 0.8160\nEpoch 229/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.2619 - accuracy: 0.9005 - val_loss: 0.5950 - val_accuracy: 0.8023\nEpoch 230/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2622 - accuracy: 0.9054 - val_loss: 0.6307 - val_accuracy: 0.8063\nEpoch 231/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2597 - accuracy: 0.9030 - val_loss: 0.6485 - val_accuracy: 0.8023\nEpoch 232/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2479 - accuracy: 0.9094 - val_loss: 0.6027 - val_accuracy: 0.8141\nEpoch 233/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.2522 - accuracy: 0.9079 - val_loss: 0.6415 - val_accuracy: 0.8043\nEpoch 234/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2564 - accuracy: 0.9054 - val_loss: 0.6362 - val_accuracy: 0.8063\nEpoch 235/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2514 - accuracy: 0.9074 - val_loss: 0.6040 - val_accuracy: 0.8004\nEpoch 236/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.2647 - accuracy: 0.9005 - val_loss: 0.6132 - val_accuracy: 0.7965\nEpoch 237/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2800 - accuracy: 0.8937 - val_loss: 0.6080 - val_accuracy: 0.7984\nEpoch 238/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.2865 - accuracy: 0.8849 - val_loss: 0.5965 - val_accuracy: 0.8023\nEpoch 239/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.2920 - accuracy: 0.8902 - val_loss: 0.5782 - val_accuracy: 0.8121\nEpoch 240/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.3139 - accuracy: 0.8780 - val_loss: 0.5731 - val_accuracy: 0.8082\nEpoch 241/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.3153 - accuracy: 0.8819 - val_loss: 0.6017 - val_accuracy: 0.8004\nEpoch 242/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2857 - accuracy: 0.8907 - val_loss: 0.6923 - val_accuracy: 0.7965\nEpoch 243/4000\n8/8 [==============================] - 1s 63ms/step - loss: 0.2789 - accuracy: 0.8966 - val_loss: 0.5590 - val_accuracy: 0.8160\nEpoch 244/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2605 - accuracy: 0.9005 - val_loss: 0.5836 - val_accuracy: 0.8043\nEpoch 245/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2398 - accuracy: 0.9074 - val_loss: 0.6317 - val_accuracy: 0.8219\nEpoch 246/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2441 - accuracy: 0.9045 - val_loss: 0.6069 - val_accuracy: 0.8317\nEpoch 247/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2445 - accuracy: 0.9064 - val_loss: 0.6772 - val_accuracy: 0.8102\nEpoch 248/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2568 - accuracy: 0.9069 - val_loss: 0.6005 - val_accuracy: 0.8219\nEpoch 249/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.2358 - accuracy: 0.9143 - val_loss: 0.6522 - val_accuracy: 0.8082\nEpoch 250/4000\n8/8 [==============================] - 1s 65ms/step - loss: 0.2311 - accuracy: 0.9192 - val_loss: 0.6418 - val_accuracy: 0.8102\nEpoch 251/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2230 - accuracy: 0.9206 - val_loss: 0.6331 - val_accuracy: 0.8200\nEpoch 252/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2092 - accuracy: 0.9260 - val_loss: 0.6534 - val_accuracy: 0.8004\nEpoch 253/4000\n8/8 [==============================] - 0s 56ms/step - loss: 0.2196 - accuracy: 0.9167 - val_loss: 0.6110 - val_accuracy: 0.8043\nEpoch 254/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2215 - accuracy: 0.9206 - val_loss: 0.6497 - val_accuracy: 0.8082\nEpoch 255/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.2119 - accuracy: 0.9299 - val_loss: 0.6268 - val_accuracy: 0.8258\nEpoch 256/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2136 - accuracy: 0.9192 - val_loss: 0.6436 - val_accuracy: 0.8102\nEpoch 257/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2137 - accuracy: 0.9216 - val_loss: 0.6256 - val_accuracy: 0.8082\nEpoch 258/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.2162 - accuracy: 0.9216 - val_loss: 0.6189 - val_accuracy: 0.8200\nEpoch 259/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2090 - accuracy: 0.9206 - val_loss: 0.6516 - val_accuracy: 0.8160\nEpoch 260/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.1972 - accuracy: 0.9309 - val_loss: 0.6799 - val_accuracy: 0.8297\nEpoch 261/4000\n8/8 [==============================] - 0s 61ms/step - loss: 0.2029 - accuracy: 0.9280 - val_loss: 0.6977 - val_accuracy: 0.8141\nEpoch 262/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2127 - accuracy: 0.9196 - val_loss: 0.7581 - val_accuracy: 0.8239\nEpoch 263/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2440 - accuracy: 0.9128 - val_loss: 0.6711 - val_accuracy: 0.8141\nEpoch 264/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2217 - accuracy: 0.9211 - val_loss: 0.6533 - val_accuracy: 0.8200\nEpoch 265/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2106 - accuracy: 0.9226 - val_loss: 0.6446 - val_accuracy: 0.8219\nEpoch 266/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2182 - accuracy: 0.9241 - val_loss: 0.6718 - val_accuracy: 0.8239\nEpoch 267/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2059 - accuracy: 0.9241 - val_loss: 0.6147 - val_accuracy: 0.8239\nEpoch 268/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2132 - accuracy: 0.9226 - val_loss: 0.6653 - val_accuracy: 0.8141\nEpoch 269/4000\n8/8 [==============================] - 0s 55ms/step - loss: 0.2430 - accuracy: 0.9079 - val_loss: 0.6761 - val_accuracy: 0.7926\nEpoch 270/4000\n8/8 [==============================] - 0s 58ms/step - loss: 0.2344 - accuracy: 0.9226 - val_loss: 0.6103 - val_accuracy: 0.8063\nEpoch 271/4000\n8/8 [==============================] - 0s 59ms/step - loss: 0.2125 - accuracy: 0.9270 - val_loss: 0.6217 - val_accuracy: 0.8239\nEpoch 272/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.1946 - accuracy: 0.9309 - val_loss: 0.6845 - val_accuracy: 0.8200\nEpoch 273/4000\n8/8 [==============================] - 0s 54ms/step - loss: 0.2044 - accuracy: 0.9241 - val_loss: 0.6918 - val_accuracy: 0.8121\nEpoch 274/4000\n8/8 [==============================] - 0s 57ms/step - loss: 0.1945 - accuracy: 0.9324 - val_loss: 0.7125 - val_accuracy: 0.8141\nEpoch 275/4000\n8/8 [==============================] - 0s 60ms/step - loss: 0.2030 - accuracy: 0.9245 - val_loss: 0.7242 - val_accuracy: 0.8219\n"
],
[
"def save_hist():\n filename = data_path + \"baseline_uci_adam256_history.csv\"\n hist_df = pd.DataFrame(hist.history) \n with open(filename, mode='w') as f:\n hist_df.to_csv(f)\nsave_hist()",
"_____no_output_____"
],
[
"fig, axes = plt.subplots(1, 2, figsize=(10, 5))\nfor i in range(1):\n ax1 = axes[0]\n ax2 = axes[1]\n\n ax1.plot(hist.history['loss'], label='training')\n ax1.plot(hist.history['val_loss'], label='validation')\n ax1.set_ylim((0.2, 1.2))\n ax1.set_title('lstm autoencoder loss')\n ax1.set_xlabel('epoch')\n ax1.set_ylabel('loss')\n ax1.legend(['train', 'validation'], loc='upper left')\n \n ax2.plot(hist.history['accuracy'], label='training')\n ax2.plot(hist.history['val_accuracy'], label='validation')\n ax2.set_ylim((0.5, 1.0))\n ax2.set_title('lstm autoencoder accuracy')\n ax2.set_xlabel('epoch')\n ax2.set_ylabel('accuracy')\n ax2.legend(['train', 'validation'], loc='upper left')\nfig.tight_layout()",
"_____no_output_____"
],
[
"def eval_model(model, ds, ds_name=\"Training\"):\n loss, acc = model.evaluate(ds, verbose=0)\n print(\"{} Dataset: loss = {} and acccuracy = {}%\".format(ds_name, np.round(loss, 3), np.round(acc*100, 2)))",
"_____no_output_____"
],
[
"eval_model(model, xtrain_ds, \"Training\")\neval_model(model, xval_ds, \"Validation\")\neval_model(model, xtest_ds, \"Test\")",
"Training Dataset: loss = 0.336 and acccuracy = 86.82%\nValidation Dataset: loss = 0.556 and acccuracy = 78.28%\nTest Dataset: loss = 0.508 and acccuracy = 80.25%\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b5fb6b847af5b0d9e7547480dfeb61d751262f | 72,548 | ipynb | Jupyter Notebook | account_summary.ipynb | mostafajoma/API_Homework | 5ad4482a94eb232ea04525e81b3912e529d0ef55 | [
"Apache-2.0"
] | null | null | null | account_summary.ipynb | mostafajoma/API_Homework | 5ad4482a94eb232ea04525e81b3912e529d0ef55 | [
"Apache-2.0"
] | null | null | null | account_summary.ipynb | mostafajoma/API_Homework | 5ad4482a94eb232ea04525e81b3912e529d0ef55 | [
"Apache-2.0"
] | 1 | 2020-07-26T21:19:27.000Z | 2020-07-26T21:19:27.000Z | 68.506138 | 32,612 | 0.756437 | [
[
[
"import os\nimport plaid\nimport requests\nimport datetime\nimport json\nimport pandas as pd\n%matplotlib inline",
"_____no_output_____"
],
[
"def pretty_print_response(response):\n print(json.dumps(response, indent=4, sort_keys=True))",
"_____no_output_____"
],
[
"PLAID_CLIENT_ID = ('PLAID_CLIENT_ID')\nPLAID_SBX_SECRET_KEY = ('PLAID_SBX_SECRET_KEY')\nPLAID_PUBLIC_KEY = ('PLAID_PUBLIC_KEY')\nPLAID_ENV = os.getenv('PLAID_ENV', 'sandbox')\nPLAID_PRODUCTS = os.getenv('PLAID_PRODUCTS', 'transactions')\n",
"_____no_output_____"
]
],
[
[
"# Plaid Access Token\n\nIn this section, you will use the plaid-python api to generate the correct authentication tokens to access data in the free developer Sandbox. This mimics how you might connect to your own account or a customer account, but due to privacy issues, this homework will only require connecting to and analyzing the fake data from the developer sandbox that Plaid provides. \n\nComplete the following steps to generate an access token:\n1. Create a client to connect to plaid\n2. Use the client to generate a public token and request the following items: \n['transactions', 'income', 'assets']\n3. Exchange the public token for an access token\n4. Test the access token by requesting and printing the available test accounts",
"_____no_output_____"
],
[
"### 1. Create a client to connect to plaid",
"_____no_output_____"
]
],
[
[
"INSTITUTION_ID = \"ins_109508\"",
"_____no_output_____"
],
[
"client = plaid.Client(client_id=PLAID_CLIENT_ID, \n secret=PLAID_SBX_SECRET_KEY, \n public_key=PLAID_PUBLIC_KEY, \n environment= PLAID_ENV )",
"_____no_output_____"
]
],
[
[
"### 2. Generate a public token",
"_____no_output_____"
]
],
[
[
"generate_tkn_response = client.Sandbox.public_token.create(INSTITUTION_ID,['transactions','income', 'assets'])\ngenerate_tkn_response",
"_____no_output_____"
]
],
[
[
"### 3. Exchange the public token for an access token",
"_____no_output_____"
]
],
[
[
"exchange_tkn_response = client.Item.public_token.exchange(generate_tkn_response['public_token'])\naccess_token = exchange_tkn_response['access_token']",
"_____no_output_____"
]
],
[
[
"### 4. Fetch Accounts",
"_____no_output_____"
]
],
[
[
"client.Accounts.get(access_token)",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Account Transactions with Plaid\n\nIn this section, you will use the Plaid Python SDK to connect to the Developer Sandbox account and grab a list of transactions. You will need to complete the following steps:\n\n\n1. Use the access token to fetch the transactions for the last 90 days\n2. Print the categories for each transaction type\n3. Create a new DataFrame using the following fields from the JSON transaction data: `date, name, amount, category`. (For categories with more than one label, just use the first category label in the list)\n4. Convert the data types to the appropriate types (i.e. datetimeindex for the date and float for the amount)",
"_____no_output_____"
],
[
"### 1. Fetch the Transactions for the last 90 days",
"_____no_output_____"
]
],
[
[
"start_date = '{:%Y-%m-%d}'.format(datetime.datetime.now() + datetime.timedelta(-90))\nend_date = '{:%Y-%m-%d}'.format(datetime.datetime.now())\ntransaction_response = client.Transactions.get(access_token,start_date,end_date)\npretty_print_response(transaction_response['transactions'][:1])",
"[\n {\n \"account_id\": \"dQA7rAZzR8HMMERWxxXwIl6k46WEvPfZzzk7A\",\n \"account_owner\": null,\n \"amount\": 25,\n \"authorized_date\": null,\n \"category\": [\n \"Payment\",\n \"Credit Card\"\n ],\n \"category_id\": \"16001000\",\n \"date\": \"2020-07-14\",\n \"iso_currency_code\": \"USD\",\n \"location\": {\n \"address\": null,\n \"city\": null,\n \"country\": null,\n \"lat\": null,\n \"lon\": null,\n \"postal_code\": null,\n \"region\": null,\n \"store_number\": null\n },\n \"merchant_name\": null,\n \"name\": \"CREDIT CARD 3333 PAYMENT *//\",\n \"payment_channel\": \"other\",\n \"payment_meta\": {\n \"by_order_of\": null,\n \"payee\": null,\n \"payer\": null,\n \"payment_method\": null,\n \"payment_processor\": null,\n \"ppd_id\": null,\n \"reason\": null,\n \"reference_number\": null\n },\n \"pending\": false,\n \"pending_transaction_id\": null,\n \"transaction_code\": null,\n \"transaction_id\": \"7rWyeWXnglIee5rE77LBHweXpZAL47IgKNQz4\",\n \"transaction_type\": \"special\",\n \"unofficial_currency_code\": null\n }\n]\n"
]
],
[
[
"### 2. Print the categories for each transaction",
"_____no_output_____"
]
],
[
[
"for transaction in transaction_response['transactions']:\n print(transaction['category'])",
"['Payment', 'Credit Card']\n['Travel', 'Taxi']\n['Transfer', 'Debit']\n['Transfer', 'Deposit']\n['Recreation', 'Gyms and Fitness Centers']\n['Travel', 'Airlines and Aviation Services']\n['Food and Drink', 'Restaurants', 'Fast Food']\n['Food and Drink', 'Restaurants', 'Coffee Shop']\n['Food and Drink', 'Restaurants']\n['Transfer', 'Credit']\n['Travel', 'Airlines and Aviation Services']\n['Travel', 'Taxi']\n['Food and Drink', 'Restaurants']\n['Payment']\n['Food and Drink', 'Restaurants', 'Fast Food']\n['Shops', 'Sporting Goods']\n['Payment', 'Credit Card']\n['Travel', 'Taxi']\n['Transfer', 'Debit']\n['Transfer', 'Deposit']\n['Recreation', 'Gyms and Fitness Centers']\n['Travel', 'Airlines and Aviation Services']\n['Food and Drink', 'Restaurants', 'Fast Food']\n['Food and Drink', 'Restaurants', 'Coffee Shop']\n['Food and Drink', 'Restaurants']\n['Transfer', 'Credit']\n['Travel', 'Airlines and Aviation Services']\n['Travel', 'Taxi']\n['Food and Drink', 'Restaurants']\n['Payment']\n['Food and Drink', 'Restaurants', 'Fast Food']\n['Shops', 'Sporting Goods']\n['Payment', 'Credit Card']\n['Travel', 'Taxi']\n['Transfer', 'Debit']\n['Transfer', 'Deposit']\n['Recreation', 'Gyms and Fitness Centers']\n['Travel', 'Airlines and Aviation Services']\n['Food and Drink', 'Restaurants', 'Fast Food']\n['Food and Drink', 'Restaurants', 'Coffee Shop']\n['Food and Drink', 'Restaurants']\n['Transfer', 'Credit']\n['Travel', 'Airlines and Aviation Services']\n['Travel', 'Taxi']\n['Food and Drink', 'Restaurants']\n['Payment']\n['Food and Drink', 'Restaurants', 'Fast Food']\n['Shops', 'Sporting Goods']\n"
]
],
[
[
"### 3. Create a new DataFrame using the following fields from the JSON transaction data: date, name, amount, category. \n\n(For categories with more than one label, just use the first category label in the list)",
"_____no_output_____"
]
],
[
[
"transaction_df = pd.DataFrame(columns=['Date','Name','Amount','Category'])\ndate = []\nname = []\namount = []\ncategory = []\nfor i in transactions:\n date.append(i['date']) \n name.append(i['name'])\n amount.append(i['amount'])\n category.append(i['category'][0])\ntransaction_df['Date'] = date\ntransaction_df['Name'] = name\ntransaction_df['Amount'] = amount\ntransaction_df['Category'] = category \ntransaction_df.head()",
"_____no_output_____"
]
],
[
[
"### 4. Convert the data types to the appropriate types \n\n(i.e. datetimeindex for the date and float for the amount)",
"_____no_output_____"
]
],
[
[
"transaction_df.dtypes",
"_____no_output_____"
],
[
"transaction_df['Date'] = pd.to_datetime(transaction_df['Date'])\ntransaction_df = transaction_df.set_index(['Date'])\ntransaction_df.dtypes",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Income Analysis with Plaid\n\nIn this section, you will use the Plaid Sandbox to complete the following:\n1. Determine the previous year's gross income and print the results\n2. Determine the current monthly income and print the results\n3. Determine the projected yearly income and print the results",
"_____no_output_____"
]
],
[
[
"income_response = client.Income.get(access_token)\npretty_print_response(income_response)",
"{\n \"income\": {\n \"income_streams\": [\n {\n \"confidence\": 0.99,\n \"days\": 720,\n \"monthly_income\": 500,\n \"name\": \"UNITED AIRLINES\"\n }\n ],\n \"last_year_income\": 6000,\n \"last_year_income_before_tax\": 7285,\n \"max_number_of_overlapping_income_streams\": 1,\n \"number_of_income_streams\": 1,\n \"projected_yearly_income\": 6085,\n \"projected_yearly_income_before_tax\": 7389\n },\n \"request_id\": \"bdOjEqQvjMgw2rh\"\n}\n"
],
[
"print(f\"Last Year's income: {income_response['income']['last_year_income_before_tax']}\")",
"Last Year's income: 7285\n"
],
[
"print(f\"Current monthly income: {income_response['income']['income_streams'][0]['monthly_income']}\")",
"Current monthly income: 500\n"
],
[
"print(f\"Projected Year's income: {income_response['income']['projected_yearly_income_before_tax']}\")",
"Projected Year's income: 7389\n"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Budget Analysis\nIn this section, you will use the transactions DataFrame to analyze the customer's budget\n\n1. Calculate the total spending per category and print the results (Hint: groupby or count transactions per category)\n2. Generate a bar chart with the number of transactions for each category \n3. Calculate the expenses per month\n4. Plot the total expenses per month",
"_____no_output_____"
],
[
"### Calculate the expenses per category",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE\nexpenses_by_category = transaction_df.groupby('Category').sum()[\"Amount\"]\nexpenses_by_category\n\n",
"_____no_output_____"
],
[
"expenses_by_category.plot(kind = \"pie\", title = \"Expenses by Category\",subplots=True, figsize = (10,10))\n",
"_____no_output_____"
]
],
[
[
"### Calculate the expenses per month",
"_____no_output_____"
]
],
[
[
"transaction_df.reset_index(inplace=True)\ntransaction_df['month'] = pd.DatetimeIndex(transaction_df['Date']).month\ntransaction_df.head()\n\n",
"_____no_output_____"
],
[
"transactions_per_month = transaction_df.groupby('month').sum()\ntransactions_per_month",
"_____no_output_____"
],
[
"transactions_per_month.plot(kind = 'bar', title = \"Transactions per Month\", rot=45)\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0b5fcc66b17bc1c1f85222fc38088660b56cd7e | 82,496 | ipynb | Jupyter Notebook | notebooks/clustering_ensemble-Copy3.ipynb | ashishlal/kaggletml | acf5d0dea3e99d12dd1795fe12bf5b83cc7e5f71 | [
"Apache-2.0"
] | null | null | null | notebooks/clustering_ensemble-Copy3.ipynb | ashishlal/kaggletml | acf5d0dea3e99d12dd1795fe12bf5b83cc7e5f71 | [
"Apache-2.0"
] | null | null | null | notebooks/clustering_ensemble-Copy3.ipynb | ashishlal/kaggletml | acf5d0dea3e99d12dd1795fe12bf5b83cc7e5f71 | [
"Apache-2.0"
] | null | null | null | 38.839925 | 1,374 | 0.455234 | [
[
[
"import numpy as np\nimport Cluster_Ensembles as CE\nfrom functools import reduce",
"_____no_output_____"
],
[
"# require(data.table)\n# require(bit64)\n# require(dbscan)\n# require(doParallel)\n# require(rBayesianOptimization)\n# path='../input/train_1/'\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport os\n\nfrom trackml.dataset import load_event, load_dataset\nfrom trackml.score import score_event\nfrom trackml.randomize import shuffle_hits\n\nfrom sklearn.preprocessing import StandardScaler\nimport hdbscan as _hdbscan\nfrom scipy import stats\nfrom tqdm import tqdm\n\nimport time\n\nfrom sklearn.cluster.dbscan_ import dbscan\nfrom sklearn.cluster import DBSCAN\nfrom sklearn.preprocessing import StandardScaler\n\nfrom sklearn.neighbors import KDTree\nimport hdbscan\nfrom bayes_opt import BayesianOptimization\n\n# https://www.ellicium.com/python-multiprocessing-pool-process/\n# http://sebastianraschka.com/Articles/2014_multiprocessing.html\nfrom multiprocessing import Pool",
"_____no_output_____"
],
[
"import os\nimport time\nimport hdbscan as _hdbscan\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\n# Input data files are available in the \"../input/\" directory.\n# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory",
"_____no_output_____"
],
[
"def create_one_event_submission(event_id, hits, labels):\n sub_data = np.column_stack(([event_id]*len(hits), hits.hit_id.values, labels))\n submission = pd.DataFrame(data=sub_data, columns=[\"event_id\", \"hit_id\", \"track_id\"]).astype(int)\n return submission",
"_____no_output_____"
],
[
"def preprocess(hits): \n x = hits.x.values\n y = hits.y.values\n z = hits.z.values\n\n r = np.sqrt(x**2 + y**2 + z**2)\n hits['x2'] = x/r\n hits['y2'] = y/r\n\n r = np.sqrt(x**2 + y**2)\n hits['z2'] = z/r\n\n ss = StandardScaler()\n X = ss.fit_transform(hits[['x2', 'y2', 'z2']].values)\n# for i, rz_scale in enumerate(self.rz_scales):\n# X[:,i] = X[:,i] * rz_scale\n\n return X",
"_____no_output_____"
],
[
"def _eliminate_outliers(clusters,M):\n my_labels = np.unique(clusters)\n norms=np.zeros((len(my_labels)),np.float32)\n indices=np.zeros((len(my_labels)),np.float32)\n for i, cluster in tqdm(enumerate(my_labels),total=len(my_labels)):\n if cluster == 0:\n continue\n index = np.argwhere(clusters==cluster)\n index = np.reshape(index,(index.shape[0]))\n indices[i] = len(index)\n x = M[index]\n norms[i] = self._test_quadric(x)\n threshold1 = np.percentile(norms,90)*5\n threshold2 = 25\n threshold3 = 6\n for i, cluster in enumerate(my_labels):\n if norms[i] > threshold1 or indices[i] > threshold2 or indices[i] < threshold3:\n clusters[clusters==cluster]=0\n \ndef _test_quadric(x):\n if x.size == 0 or len(x.shape)<2:\n return 0\n Z = np.zeros((x.shape[0],10), np.float32)\n Z[:,0] = x[:,0]**2\n Z[:,1] = 2*x[:,0]*x[:,1]\n Z[:,2] = 2*x[:,0]*x[:,2]\n Z[:,3] = 2*x[:,0]\n Z[:,4] = x[:,1]**2\n Z[:,5] = 2*x[:,1]*x[:,2]\n Z[:,6] = 2*x[:,1]\n Z[:,7] = x[:,2]**2\n Z[:,8] = 2*x[:,2]\n Z[:,9] = 1\n v, s, t = np.linalg.svd(Z,full_matrices=False) \n smallest_index = np.argmin(np.array(s))\n T = np.array(t)\n T = T[smallest_index,:] \n norm = np.linalg.norm(np.dot(Z,T), ord=2)**2\n return norm",
"_____no_output_____"
],
[
"# Input data files are available in the \"../input/\" directory.\n# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory\n\n#------------------------------------------------------\n\ndef make_counts(labels):\n \n \n _,reverse,count = np.unique(labels,return_counts=True,return_inverse=True)\n counts = count[reverse]\n counts[labels==0]=0\n \n return counts\n\ndef one_loop(param):\n \n # <todo> tune your parameters or design your own features here! \n \n i,m, x,y,z, d,r, a, a_start,a_step = param\n #print('\\r %3d %+0.8f '%(i,da), end='', flush=True)\n \n da = m*(a_start - (i*a_step))\n aa = a + np.sign(z)*z*da \n zr = z/r\n \n X = StandardScaler().fit_transform(np.column_stack([aa, aa/zr, zr, 1/zr, aa/zr + 1/zr]))\n _,l = dbscan(X, eps=0.0035, min_samples=1,)\n\n return l\n\ndef one_loop1(param):\n \n # <todo> tune your parameters or design your own features here! \n \n i,m, x,y,z, d,r,r2,z2,a, a_start,a_step = param\n #print('\\r %3d %+0.8f '%(i,da), end='', flush=True)\n \n da = m*(a_start - (i*a_step))\n aa = a + np.sign(z)*z*da\n# if m == 1:\n# print(da)\n zr = z/r # this is cot(theta), 1/zr is tan(theta)\n theta = np.arctan2(r, z)\n ct = np.cos(theta)\n st = np.sin(theta)\n tt = np.tan(theta)\n# ctt = np.cot(theta)\n z2r = z2/r\n z2r2 = z2/r2\n# X = StandardScaler().fit_transform(df[['r2', 'theta_1', 'dip_angle', 'z2', 'z2_1', 'z2_2']].values)\n\n caa = np.cos(aa)\n saa = np.sin(aa)\n taa = np.tan(aa)\n ctaa = 1/taa\n \n# 0.000005\n deps = 0.0000025\n X = StandardScaler().fit_transform(np.column_stack([caa, saa, tt, 1/tt]))\n l= DBSCAN(eps=0.0035+i*deps,min_samples=1,metric='euclidean',n_jobs=8).fit(X).labels_\n# _,l = dbscan(X, eps=0.0035, min_samples=1,algorithm='auto')\n \n return l\n\ndef one_loop2(param):\n \n # <todo> tune your parameters or design your own features here! \n \n i,m, x,y,z, d,r,r2,z2,a, a_start,a_step = param\n #print('\\r %3d %+0.8f '%(i,da), end='', flush=True)\n \n da = m*(a_start - (i*a_step))\n aa = a + np.sign(z)*z*da\n# if m == 1:\n# print(da)\n zr = z/r # this is cot(theta), 1/zr is tan(theta)\n theta = np.arctan2(r, z)\n ct = np.cos(theta)\n st = np.sin(theta)\n tt = np.tan(theta)\n# ctt = np.cot(theta)\n z2r = z2/r\n z2r2 = z2/r2\n# X = StandardScaler().fit_transform(df[['r2', 'theta_1', 'dip_angle', 'z2', 'z2_1', 'z2_2']].values)\n\n caa = np.cos(aa)\n saa = np.sin(aa)\n taa = np.tan(aa)\n ctaa = 1/taa\n \n# 0.000005\n deps = 0.0000025\n X = StandardScaler().fit_transform(np.column_stack([caa, saa, tt, 1/tt]))\n l= DBSCAN(eps=0.0035+i*deps,min_samples=1,metric='euclidean',n_jobs=8).fit(X).labels_\n# _,l = dbscan(X, eps=0.0035, min_samples=1,algorithm='auto')\n \n return l\n\ndef do_dbscan_predict(df):\n \n x = df.x.values\n y = df.y.values\n z = df.z.values\n r = np.sqrt(x**2+y**2)\n d = np.sqrt(x**2+y**2+z**2)\n a = np.arctan2(y,x)\n\n x2 = df['x']/d\n y2 = df['y']/d\n z2 = df['z']/r\n\n r2 = np.sqrt(x2**2 + y2**2)\n phi = np.arctan2(y, x)\n phi_deg= np.degrees(np.arctan2(y, x))\n phi2 = np.arctan2(y2, x2)\n phi2_deg = np.degrees(np.arctan2(y2, x2))\n \n for angle in range(-180,180,1):\n \n df1 = df.loc[(df.phi_deg>(angle-1.0)) & (df.phi_deg<(angle+1.0))]\n \n x = df1.x.values\n y = df1.y.values\n z = df1.z.values\n r = np.sqrt(x**2+y**2)\n d = np.sqrt(x**2+y**2+z**2)\n a = np.arctan2(y,x)\n\n x2 = df1['x']/d\n y2 = df1['y']/d\n z2 = df1['z']/r\n \n r2 = np.sqrt(x2**2 + y2**2)\n theta= np.arctan2(r, z)\n theta1 = np.arctan2(r2, z2)\n\n \n tan_dip = phi/theta\n tan_dip1 = phi/z2\n z2_1 = 1/z2\n z2_2 = phi/z2 + 1/z2\n\n dip_angle = np.arctan2(z2, (np.sqrt(x2**2 +y2**2)) * np.arccos(x2/np.sqrt(x2**2 + y2**2)))\n dip_angle1 = np.arctan2(z, (np.sqrt(x**2 +y**2)) * np.arccos(x2/np.sqrt(x**2 + y**2)))\n scores = []\n\n a_start,a_step,a_num = 0.00100,0.0000095,150\n \n params = [(i,m, x,y,z,d,r,r2,z2, a, a_start,a_step) for i in range(a_num) for m in [-1,1]]\n\n if 1: \n pool = Pool(processes=1)\n ls = pool.map( one_loop1, params )\n\n\n if 0:\n ls = [ one_loop(param) for param in params ]\n\n\n ##------------------------------------------------\n\n num_hits=len(df)\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n for l in ls:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n \n\n# cl = hdbscan.HDBSCAN(min_samples=1,min_cluster_size=7,\n# metric='braycurtis',cluster_selection_method='leaf',algorithm='best', \n# leaf_size=50)\n \n# X = preprocess(df)\n# l1 = pd.Series(labels)\n# labels = np.unique(l1)\n \n# # print(X.shape)\n# # print(len(labels_org))\n# # print(len(labels_org[labels_org ==0]))\n# # print(len(labels_org[labels_org ==-1]))\n \n# n_labels = 0\n# while n_labels < len(labels):\n# n_labels = len(labels)\n# max_len = np.max(l1)\n# s = list(l1[l1 == 0].keys())\n# X = X[s]\n# print(X.shape)\n# if X.shape[0] <= 1:\n# break\n# l = cl.fit_predict(X)+max_len\n# # print(len(l))\n# l1[l1 == 0] = l\n# labels = np.unique(l1)\n \n return labels\n\n## reference----------------------------------------------\ndef do_dbscan0_predict(df):\n x = df.x.values\n y = df.y.values\n z = df.z.values\n r = np.sqrt(x**2+y**2)\n d = np.sqrt(x**2+y**2+z**2)\n\n X = StandardScaler().fit_transform(np.column_stack([\n x/d, y/d, z/r]))\n _,labels = dbscan(X,\n eps=0.0075,\n min_samples=1,\n algorithm='auto',\n n_jobs=-1)\n\n #labels = hdbscan(X, min_samples=1, min_cluster_size=5, cluster_selection_method='eom')\n\n return labels\n\n## reference----------------------------------------------\ndef do_dbscan0_predict(df):\n x = df.x.values\n y = df.y.values\n z = df.z.values\n r = np.sqrt(x**2+y**2)\n d = np.sqrt(x**2+y**2+z**2)\n\n X = StandardScaler().fit_transform(np.column_stack([\n x/d, y/d, z/r]))\n _,labels = dbscan(X,\n eps=0.0075,\n min_samples=1,\n algorithm='auto',\n n_jobs=-1)\n\n #labels = hdbscan(X, min_samples=1, min_cluster_size=5, cluster_selection_method='eom')\n\n return labels\n\ndef extend(submission,hits):\n df = submission.merge(hits, on=['hit_id'], how='left')\n# df = submission.append(hits)\n# print(df.head())\n df = df.assign(d = np.sqrt( df.x**2 + df.y**2 + df.z**2 ))\n df = df.assign(r = np.sqrt( df.x**2 + df.y**2))\n df = df.assign(arctan2 = np.arctan2(df.z, df.r))\n\n for angle in range(-180,180,1):\n\n print ('\\r %f'%angle, end='',flush=True)\n #df1 = df.loc[(df.arctan2>(angle-0.5)/180*np.pi) & (df.arctan2<(angle+0.5)/180*np.pi)]\n df1 = df.loc[(df.arctan2>(angle-1.0)/180*np.pi) & (df.arctan2<(angle+1.0)/180*np.pi)]\n\n min_num_neighbours = len(df1)\n if min_num_neighbours<4: continue\n\n hit_ids = df1.hit_id.values\n x,y,z = df1.as_matrix(columns=['x', 'y', 'z']).T\n r = (x**2 + y**2)**0.5\n r = r/1000\n a = np.arctan2(y,x)\n tree = KDTree(np.column_stack([a,r]), metric='euclidean')\n\n track_ids = list(df1.track_id.unique())\n num_track_ids = len(track_ids)\n min_length=3\n\n for i in range(num_track_ids):\n p = track_ids[i]\n if p==0: continue\n\n idx = np.where(df1.track_id==p)[0]\n if len(idx)<min_length: continue\n\n if angle>0:\n idx = idx[np.argsort( z[idx])]\n else:\n idx = idx[np.argsort(-z[idx])]\n\n\n ## start and end points ##\n idx0,idx1 = idx[0],idx[-1]\n a0 = a[idx0]\n a1 = a[idx1]\n r0 = r[idx0]\n r1 = r[idx1]\n\n da0 = a[idx[1]] - a[idx[0]] #direction\n dr0 = r[idx[1]] - r[idx[0]]\n direction0 = np.arctan2(dr0,da0) \n\n da1 = a[idx[-1]] - a[idx[-2]]\n dr1 = r[idx[-1]] - r[idx[-2]]\n direction1 = np.arctan2(dr1,da1) \n\n\n ## extend start point\n ns = tree.query([[a0,r0]], k=min(20,min_num_neighbours), return_distance=False)\n ns = np.concatenate(ns)\n direction = np.arctan2(r0-r[ns],a0-a[ns])\n ns = ns[(r0-r[ns]>0.01) &(np.fabs(direction-direction0)<0.04)]\n\n for n in ns:\n df.loc[ df.hit_id==hit_ids[n],'track_id' ] = p \n\n ## extend end point\n ns = tree.query([[a1,r1]], k=min(20,min_num_neighbours), return_distance=False)\n ns = np.concatenate(ns)\n\n direction = np.arctan2(r[ns]-r1,a[ns]-a1)\n ns = ns[(r[ns]-r1>0.01) &(np.fabs(direction-direction1)<0.04)] \n\n for n in ns:\n df.loc[ df.hit_id==hit_ids[n],'track_id' ] = p\n #print ('\\r')\n# df = df[['particle_id', 'weight', 'event_id', 'hit_id', 'track_id']]\n df = df[['event_id', 'hit_id', 'track_id']]\n return df",
"_____no_output_____"
],
[
"import hdbscan\nimport math\nseed = 123\nnp.random.seed(seed)\n\ndef shift(l, n):\n return l[n:] + l[:n]\n\n# https://stackoverflow.com/questions/29246455/python-setting-decimal-place-range-without-rounding\ndef truncate(f, n):\n return math.floor(f * 10 ** n) / 10 ** n\n\ndef trackML31(df, w1, w2, w3, w4, w5, w6, w7, w8, w9, w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_shift, r0Inv_nu):\n x = df.x.values\n y = df.y.values\n z = df.z.values\n \n# dz = 0\n z = z + z_shift\n \n rt = np.sqrt(x**2+y**2)\n r = np.sqrt(x**2+y**2+z**2)\n a0 = np.arctan2(y,x)\n\n x2 = x/r\n y2 = y/r\n \n \n phi = np.arctan2(y, x)\n phi_deg= np.degrees(np.arctan2(y, x))\n \n \n z1 = z/rt\n z2 = z/r\n \n z3 = np.log1p(abs(z/r))*np.sign(z)\n \n x1 = x/rt\n y1 = y/rt\n \n y3 = np.log1p(abs(y/r))*np.sign(y)\n \n theta = np.arctan2(rt, z)\n theta_deg = np.degrees(np.arctan2(rt, z))\n tt = np.tan(theta_deg)\n \n z4 = np.sqrt(abs(z/rt))\n \n x4 = np.sqrt(abs(x/r))\n y4 = np.sqrt(abs(y/r))\n \n mm = 1\n ls = []\n \n\n# def f(x):\n# return a0+mm*(rt+ 0.0000145*rt**2)/1000*(x/2)/180*np.pi\n \n for ii in range(Niter):\n \n mm = mm * (-1)\n\n a1 = a0+mm*(rt+ 0.0000145*rt**2)/1000*(ii/2)/180*np.pi\n \n da1 = mm*(1 + (2 * 0.0000145 * rt))/1000*(ii/2)/180*np.pi\n ia1 = a0*rt + mm*(((rt**2)/2) + (0.0000145*rt**3)/3)/1000*(ii/2)/180*np.pi\n \n saa = np.sin(a1)\n caa = np.cos(a1)\n \n raa = x*caa + y*saa\n \n t1 = theta+mm*(rt+ 0.8435*rt**2)/1000*(ii/2)/180*np.pi\n ctt = np.cos(t1)\n stt = np.sin(t1)\n \n# mom = np.sqrt(1 + (z1 **2))\n# mom2 = r0Inv * np.sqrt(1 + (z2 **2)).round(2)\n mom2 = [truncate(np.sqrt(1 + (i**2)),4) for i in z2]\n \n# theta0= np.arcsin[np.sqrt(x**2+y**2)/(2*R)]- a0\n r0_list = list(np.linspace(30, 3100, 100))\n for r0 in r0_list:\n r0Inv = 1./r0\n theta0= np.nan_to_num(np.arcsin(rt*0.5*r0Inv))- a0\n# r0Inv = 2. * np.cos(a0 - theta0) / rt\n \n# r0Inv = 2. * caa / r\n# r0Inv2 = 2. * np.cos(a0 - theta0) / rt\n# r0Inv_d = -2. * np.sin(a1-t1) * da1 /r\n # https://www.kaggle.com/okhlopkov/attempts-to-struggle-with-clustering\n# fundu = np.arcsin((y * np.sin(theta0) - x * np.cos(theta0)) / rt) / (z - z0)\n# fundu = np.arcsin((y * np.sin(theta0) - x * np.cos(theta0)) / rt) / (z-z_shift)\n# fundu2 = np.arcsin((y * np.sin(theta0) - x * np.cos(theta0)) / r) / (z-z_shift)\n X = StandardScaler().fit_transform(np.column_stack([caa, saa, z1, z2, rt/r, x/r, y/r, z3, y1, y3, \n z4, x4, y4, raa, mom2, \n da1, ia1, theta0]))\n\n # X = StandardScaler().fit_transform(np.column_stack([caa, saa, z1, z2, fundu2]))\n # print(X.shape)\n\n # X = StandardScaler().fit_transform(np.column_stack([caa,saa,z1,z2]))\n\n cx = [w1,w1,w2,w3, w4, w5, w6, w7, w8, w9, w11, w12, w13, w14, w15, w16, w17, 1.]\n\n # cx = [w1,w1,w2,w3, w15]\n\n X = np.multiply(X, cx)\n\n l= DBSCAN(eps=0.004,min_samples=1,metric='euclidean',n_jobs=4).fit(X).labels_\n\n ls.append(l)\n \n \n num_hits=len(df)\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n lss = []\n for l in ls:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n \n# lss.append(labels)\n \n# for i in range(Niter):\n# labels1 = np.zeros(num_hits,np.int32)\n# counts1 = np.zeros(num_hits,np.int32)\n# ls1 = ls.copy()\n# ls1 = shift(ls1, 1)\n# np.random.shuffle(ls1)\n# for l in ls1:\n# c = make_counts(l)\n# idx = np.where((c-counts>0) & (c<20))[0]\n# labels1[idx] = l[idx] + labels1.max()\n# counts1 = make_counts(labels1)\n# l1 = labels1.copy()\n# lss.append(l1)\n\n\n# labels = np.zeros(num_hits,np.int32)\n# counts = np.zeros(num_hits,np.int32)\n \n# for l in lss:\n# c = make_counts(l)\n# idx = np.where((c-counts>0) & (c<20))[0]\n# labels[idx] = l[idx] + labels.max()\n# counts = make_counts(labels)\n \n# df = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id', 'vlm'],\n# data=np.column_stack(([int(0),]*len(hits), hits.hit_id.values, labels, hits.vlm.values))\n# )\n# df = pd.DataFrame()\n# df['hit_id']=hits.hit_id.values\n# df['vlm'] = hits.vlm.values\n# df['track_id'] = labels\n \n# for l in np.unique(labels):\n# df_l = df[df.track_id == l]\n# df_l['vlm_count'] =df_l.groupby('vlm')['vlm'].transform('count')\n# same_vlm_multiple_hits = np.any(df_l.vlm_count > 1)\n# if same_vlm_multiple_hits == True:\n# print(l)\n# which_vlm_multiple_hits = list(df_l[df_l.vlm_count > 1].index) \n# which_vlm_multiple_hits.pop(0)\n \n# df.loc[which_vlm_multiple_hits, 'track_id'] = 9999999999\n \n# return df.track_id.values\n# sub = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n# data=np.column_stack(([int(0),]*len(df), df.hit_id.values, labels))\n# )\n# sub['track_count'] = sub.groupby('track_id')['track_id'].transform('count')\n# # sub.loc[sub.track_count < 5, 'track_id'] = 0\n \n# sub1 = sub[sub.track_id < 0]\n# sub2 = sub[sub.track_id >= 0]\n# L_neg = sub1.track_id.values\n# L_pos = sub2.track_id.values\n# a = 1\n# for l in L_neg:\n# for l1 in range(a, np.iinfo(np.int32).max):\n# if l1 in L_pos:\n# continue\n# sub.loc[sub.track_id == l, 'track_id'] = l1\n# a = l1 +1\n# break\n \n# L = list(sub.track_id.values)\n# labels = np.zeros(num_hits,np.int32)\n# for ii in range(num_hits):\n# labels[ii] = L[ii]\n# print(np.any(labels < 0))\n return labels",
"_____no_output_____"
],
[
"%%time\n# def run_dbscan():\n\ndata_dir = '../data/train'\n\n# event_ids = [\n# '000001030',##\n# '000001025','000001026','000001027','000001028','000001029',\n# ]\n\nevent_ids = [\n '000001030',##\n\n]\n\nsum=0\nsum_score=0\nfor i,event_id in enumerate(event_ids):\n particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)\n hits = pd.read_csv(data_dir + '/event%s-hits.csv'%event_id)\n cells = pd.read_csv(data_dir + '/event%s-cells.csv'%event_id)\n truth = pd.read_csv(data_dir + '/event%s-truth.csv'%event_id)\n particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)\n \n truth = pd.merge(truth, particles, how='left', on='particle_id')\n hits = pd.merge(hits, truth, how='left', on='hit_id')\n ",
"CPU times: user 3.72 s, sys: 392 ms, total: 4.11 s\nWall time: 1.79 s\n"
],
[
"%%time\n\nw1 = 1.1932215111905984\nw2 = 0.39740553885387364\nw3 = 0.3512647720585538\nw4 = 0.1470\nw5 = 0.01201\nw6 = 0.0003864\nw7 = 0.0205\nw8 = 0.0049\nw9 = 0.00121\nw10 = 1.4930496676654575e-05\nw11 = 0.0318\nw12 = 0.000435\nw13 = 0.00038\nw14 = 0.00072\nw15 = 5.5e-05\n# w15 = 0.000265\nw16 = 0.0031\nw17 = 0.00021\nw18 = 7.5e-05\n\nNiter=247 \nprint(w18)\n\nz_shift = 0\n# ls = []\ntrack_id = trackML31(hits, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_shift)\n\nsum_score=0\nsum = 0\nsubmission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(event_id),]*len(hits), hits.hit_id.values, track_id))\n).astype(int)\n\n\nfor i in range(8):\n submission = extend(submission,hits)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\nprint('--------------------------------------')\nsc = sum_score/sum\nprint(sc)\n# caa, saa: 5 mins score 0\n# caa, saa, z1: 0.3942327679531816, 6 min 14s\n# z1: 5.99028402551861e-05, 11 mins\n# caa,saa,z1,z2: 7 mins, 0.5315668141457246",
"7.5e-05\n 179.0000000[ 0] score : 0.52615557\n 179.0000000[ 1] score : 0.53028442\n 179.0000000[ 2] score : 0.53194035\n 179.0000000[ 3] score : 0.53245126\n 179.0000000[ 4] score : 0.53269416\n 179.0000000[ 5] score : 0.53334090\n 179.0000000[ 6] score : 0.53274646\n 179.0000000[ 7] score : 0.53292140\n--------------------------------------\n0.5315668141457246\nCPU times: user 6min 52s, sys: 4.24 s, total: 6min 57s\nWall time: 6min 54s\n"
],
[
"num_hits = len(hits)\nlabels = np.zeros(num_hits,np.int32)\ncounts = np.zeros(num_hits,np.int32)\nfor i in range(len(ls)):\n labels1 = np.zeros(num_hits,np.int32)\n counts1 = np.zeros(num_hits,np.int32)\n ls1 = ls.copy()\n ls1 = shift(ls1, 1)\n np.random.shuffle(ls1)\n for l in ls1:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels1[idx] = l[idx] + labels1.max()\n counts1 = make_counts(labels1)\n l1 = labels1.copy()\n lss.append(l1)\n\n\nlabels = np.zeros(num_hits,np.int32)\ncounts = np.zeros(num_hits,np.int32)\n\nfor l in lss:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n\nsum_score=0\nsum = 0\nsubmission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(event_id),]*len(hits), hits.hit_id.values, labels))\n).astype(int)\n\n\nfor i in range(8):\n submission = extend(submission,hits)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\nprint('--------------------------------------')\nsc = sum_score/sum\nprint(sc)\n\n# 179.0000000[ 0] score : 0.63363358\n# 179.0000000[ 1] score : 0.63765912\n# 179.0000000[ 2] score : 0.63883962\n# 179.0000000[ 3] score : 0.64030808\n# 179.0000000[ 4] score : 0.64120567\n# 179.0000000[ 5] score : 0.64168075\n# 179.0000000[ 6] score : 0.64064708\n# 179.0000000[ 7] score : 0.64116239\n# --------------------------------------\n# 0.63939203643381",
" 179.0000000[ 0] score : 0.63363358\n 179.0000000[ 1] score : 0.63765912\n 179.0000000[ 2] score : 0.63883962\n 179.0000000[ 3] score : 0.64030808\n 179.0000000[ 4] score : 0.64120567\n 179.0000000[ 5] score : 0.64168075\n 179.0000000[ 6] score : 0.64064708\n 179.0000000[ 7] score : 0.64116239\n--------------------------------------\n0.63939203643381\n"
],
[
"hits.head()",
"_____no_output_____"
],
[
"def Fun4BO22221(params):\n tic = t = time.time()\n df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, z_s, t = params\n \n l = trackML31(df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_s, t)\n \n toc = time.time()\n print('\\r z_s : %0.6f, t: %0.6f , %0.0f min'%(z_s, t, (toc-tic)/60))\n\n return l",
"_____no_output_____"
],
[
"%%time\nls = []\nlss = []\ndef Fun4BO21(df, truth):\n \n w1 = 1.1932215111905984\n w2 = 0.39740553885387364\n w3 = 0.3512647720585538\n w4 = 0.1470\n w5 = 0.01201\n w6 = 0.0003864\n w7 = 0.0205\n w8 = 0.0049\n w9 = 0.00121\n w10 = 1.4930496676654575e-05\n w11 = 0.0318\n w12 = 0.000435\n w13 = 0.00038\n w14 = 0.00072\n w15 = 0.01\n# w15 = 0.00109\n# w15 = 0.001\n# w15 = 5.5e-05\n\n# w15 = 1.\n w16 = 0.0031\n w17 = 0.00021\n w18 = 7.5e-05\n \n Niter=247 \n# print(w18)\n \n \n# z_shift = 0\n T = []\n L = []\n params = []\n if 1:\n L = ['r0Inv']\n params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n 0, 0))\n pool = Pool(processes=1)\n ls1 = pool.map(Fun4BO22221, params, chunksize=1)\n pool.close()\n \n j = 0\n ls = ls1\n num_hits = len(df)\n # lss = []\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n\n ls2 = []\n if 1:\n for l in ls:\n print(L[j])\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))\n ).astype(int)\n\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(j, score))\n \n for i in range(8):\n submission = extend(submission,df)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n# l2 = submission.track_id.values\n# ls2.append(l2)\n j += 1\n if 0:\n L = [1., 0.1,0.01, 0.001, 0.0001]\n for w15 in L:\n params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n 0, 0))\n pool = Pool(processes=len(L))\n ls1 = pool.map(Fun4BO22221, params, chunksize=1)\n pool.close()\n \n j = 0\n ls = ls1\n num_hits = len(df)\n # lss = []\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n\n ls2 = []\n if 1:\n for l in ls:\n print(L[j])\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))\n ).astype(int)\n\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(j, score))\n \n for i in range(8):\n submission = extend(submission,df)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n# l2 = submission.track_id.values\n# ls2.append(l2)\n j += 1\n if 0:\n params = []\n r0Inv = [1]\n r0Inv_list = list(np.linspace(-180, 180, 10))\n\n theta0 = theta0 + theta_list\n z_shifts = [0]\n z_list = list(np.linspace(-5.5, 5.5, 10))\n z_shifts = z_shifts + z_list\n for z_s in z_shifts:\n t = 0\n if z_s != 0:\n np.random.shuffle(theta0)\n t = theta0.pop()\n T.append(t)\n L.append(z_s)\n params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_s, t))\n \n pool = Pool(processes=len(z_shifts))\n ls1 = pool.map(Fun4BO22221, params, chunksize=1)\n pool.close()\n \n j = 0\n ls = ls1\n num_hits = len(df)\n # lss = []\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n ls2 = []\n if 0:\n for l in ls:\n print(L[j], T[j])\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))\n ).astype(int)\n\n score = score_event(truth, submission)\n print('[%2d, %2d] score : %0.8f'%(L[j], T[j], score))\n \n for i in range(8):\n submission = extend(submission,df)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n l2 = submission.track_id.values\n ls2.append(l2)\n j += 1\n \n# labels = np.zeros(num_hits,np.int32)\n# counts = np.zeros(num_hits,np.int32)\n for i in range(len(ls2)): \n labels1 = np.zeros(num_hits,np.int32)\n counts1 = np.zeros(num_hits,np.int32)\n ls1 = ls.copy()\n ls1 = shift(ls1, 1)\n np.random.shuffle(ls1)\n for l in ls1:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels1[idx] = l[idx] + labels1.max()\n counts1 = make_counts(labels1)\n l1 = labels1.copy()\n lss.append(l1)\n \n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n\n for l in lss:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(0),]*len(df), df.hit_id.values, labels))\n ).astype(int)\n\n\n for i in range(8):\n submission = extend(submission,df)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n if 0:\n params = []\n theta0 = [0]\n theta_list = list(np.linspace(-180, 180, 10))\n\n theta0 = theta0 + theta_list\n z_shifts = [0]\n z_list = list(np.linspace(-5.5, 5.5, 10))\n z_shifts = z_shifts + z_list\n for z_s in z_shifts:\n t = 0\n if z_s != 0:\n np.random.shuffle(theta0)\n t = theta0.pop()\n T.append(t)\n L.append(z_s)\n params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_s, t))\n \n pool = Pool(processes=len(z_shifts))\n ls1 = pool.map(Fun4BO22221, params, chunksize=1)\n pool.close()\n \n j = 0\n ls = ls1\n num_hits = len(df)\n # lss = []\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n ls2 = []\n if 0:\n for l in ls:\n print(L[j], T[j])\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(0),]*len(df), df.hit_id.values, l))\n ).astype(int)\n\n score = score_event(truth, submission)\n print('[%2d, %2d] score : %0.8f'%(L[j], T[j], score))\n \n for i in range(8):\n submission = extend(submission,df)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n l2 = submission.track_id.values\n ls2.append(l2)\n j += 1\n \n# labels = np.zeros(num_hits,np.int32)\n# counts = np.zeros(num_hits,np.int32)\n for i in range(len(ls2)): \n labels1 = np.zeros(num_hits,np.int32)\n counts1 = np.zeros(num_hits,np.int32)\n ls1 = ls.copy()\n ls1 = shift(ls1, 1)\n np.random.shuffle(ls1)\n for l in ls1:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels1[idx] = l[idx] + labels1.max()\n counts1 = make_counts(labels1)\n l1 = labels1.copy()\n lss.append(l1)\n \n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n\n for l in lss:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(0),]*len(df), df.hit_id.values, labels))\n ).astype(int)\n\n\n for i in range(8):\n submission = extend(submission,df)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n \n if 0:\n theta0 = [0]\n theta_list = list(np.linspace(-np.pi, np.pi, 50))\n\n theta0 = theta0 + theta_list\n params = []\n for t in theta0:\n params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n 0, t))\n\n pool = Pool(processes=20)\n ls1 = pool.map(Fun4BO22221, params, chunksize=1)\n pool.close()\n\n ls = ls + ls1\n num_hits = len(df)\n # lss = []\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n for i in range(len(ls)):\n labels1 = np.zeros(num_hits,np.int32)\n counts1 = np.zeros(num_hits,np.int32)\n ls1 = ls.copy()\n ls1 = shift(ls1, 1)\n np.random.shuffle(ls1)\n for l in ls1:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels1[idx] = l[idx] + labels1.max()\n counts1 = make_counts(labels1)\n l1 = labels1.copy()\n lss.append(l1)\n\n\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n\n for l in lss:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n\n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(event_id),]*len(df), df.hit_id.values, labels))\n ).astype(int)\n\n\n for i in range(8):\n submission = extend(submission,hits)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n \n return sc\n return 0\n# return labels",
"CPU times: user 0 ns, sys: 0 ns, total: 0 ns\nWall time: 83.9 µs\n"
],
[
"def Fun4BO2222(params):\n df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, z_s = params\n \n l = trackML31(df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_s)\n return l",
"_____no_output_____"
],
[
"%%time\n\ndef Fun4BO2(df):\n \n w1 = 1.1932215111905984\n w2 = 0.39740553885387364\n w3 = 0.3512647720585538\n w4 = 0.1470\n w5 = 0.01201\n w6 = 0.0003864\n w7 = 0.0205\n w8 = 0.0049\n w9 = 0.00121\n w10 = 1.4930496676654575e-05\n w11 = 0.0318\n w12 = 0.000435\n w13 = 0.00038\n w14 = 0.00072\n w15 = 0.00109\n \n# w15 = 5.5e-05\n# w15 = 0.000265\n w16 = 0.0031\n w17 = 0.00021\n w18 = 7.5e-05\n \n Niter=247 \n# print(w18)\n \n ls = []\n lss = []\n z_shift = 0\n\n z_shifts = [0]\n z_shift_list = list(np.linspace(-5.5, 5.5, 5))\n \n z_shifts = z_shifts + z_shift_list\n params = []\n for z_s in z_shifts:\n params.append((df, w1,w2,w3,w4,w5,w6,w7,w8,w9,w10, w11, w12, w13, w14, w15, w16, w17, w18, Niter, \n z_s))\n \n pool = Pool(processes=6)\n ls1 = pool.map(Fun4BO2222, params, chunksize=1)\n pool.close()\n \n ls = ls + ls1\n num_hits = len(df)\n# lss = []\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n for i in range(len(ls)):\n labels1 = np.zeros(num_hits,np.int32)\n counts1 = np.zeros(num_hits,np.int32)\n ls1 = ls.copy()\n ls1 = shift(ls1, 1)\n np.random.shuffle(ls1)\n for l in ls1:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels1[idx] = l[idx] + labels1.max()\n counts1 = make_counts(labels1)\n l1 = labels1.copy()\n lss.append(l1)\n\n\n labels = np.zeros(num_hits,np.int32)\n counts = np.zeros(num_hits,np.int32)\n \n for l in lss:\n c = make_counts(l)\n idx = np.where((c-counts>0) & (c<20))[0]\n labels[idx] = l[idx] + labels.max()\n counts = make_counts(labels)\n \n sum_score=0\n sum = 0\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([int(event_id),]*len(df), df.hit_id.values, labels))\n ).astype(int)\n \n \n for i in range(8):\n submission = extend(submission,hits)\n score = score_event(truth, submission)\n print('[%2d] score : %0.8f'%(i, score))\n sum_score += score\n sum += 1\n\n print('--------------------------------------')\n sc = sum_score/sum\n print(sc)\n \n return sc\n# return labels",
"CPU times: user 0 ns, sys: 0 ns, total: 0 ns\nWall time: 41.2 µs\n"
],
[
"%%time\n# def run_dbscan():\n\ndata_dir = '../data/train'\n\n# event_ids = [\n# '000001030',##\n# '000001025','000001026','000001027','000001028','000001029',\n# ]\n\nevent_ids = [\n '000001030',##\n\n]\n\nsum=0\nsum_score=0\nfor i,event_id in enumerate(event_ids):\n particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)\n hits = pd.read_csv(data_dir + '/event%s-hits.csv'%event_id)\n cells = pd.read_csv(data_dir + '/event%s-cells.csv'%event_id)\n truth = pd.read_csv(data_dir + '/event%s-truth.csv'%event_id)\n particles = pd.read_csv(data_dir + '/event%s-particles.csv'%event_id)\n \n truth = pd.merge(truth, particles, how='left', on='particle_id')\n hits = pd.merge(hits, truth, how='left', on='hit_id')\n \n# bo = BayesianOptimization(Fun4BO,pbounds = {'w1':w1,'w2':w2,'w3':w3,'Niter':Niter})\n# bo.maximize(init_points = 3, n_iter = 20, acq = \"ucb\", kappa = 2.576)\n# w1 = 1.1932215111905984\n# w2 = 0.39740553885387364\n# w3 = 0.3512647720585538\n# w4 = [0.1, 0.2] # 0.1470 -> 0.55690\n# w4 = 0.1470\n# w5 = [0.001, 1.2] # 0.7781 -> 0.55646, 0.7235 + N = 247 => 0.56025\n \n# Niter = 179\n# Niter = 247\n# w5 = 0.01\n# for w6 in [0.012, 0.01201, 0.01202, 0.01203, 0.01204, 0.01205, 0.01206, 0.01207, 0.01208, 0.01209, 0.0121]:\n# EPS = 1e-12\n# w6 = [0.001, 1.2]\n# w6 = 0.0205\n# w18 = [0.00001, 0.05]\n# w13 = 0.00038\n# w14 = 0.0007133505234834969\n# for w8 in np.arange(0.00008, 0.00015, 0.000005):\n# print(w8)\n# Fun4BO2(1)\n# for w18 in np.arange(1.0e-05, 9.0e-05, 5.0e-06):\n# print(w18)\n Fun4BO21(hits, truth)\n# Niter = [240, 480]\n# w18 = [0.00001, 0.0003]\n# bo = BayesianOptimization(Fun4BO2,pbounds = {'w18':w18})\n# bo.maximize(init_points = 20, n_iter = 5, acq = \"ucb\", kappa = 2.576)\n\n# x/y: 7 | 06m30s | 0.55302 | 0.0100 | \n# x/y: 0.001: 0.55949\n# x/y: 0.0001: 0.55949\n# x/y: 0.002: 0.55959\n# x/y: 0.003: 0.55915\n# x/y: 0.0025: 0.55925\n# x/y: 0.0015: 0.55953\n# x/r: 0.0015: 0.56186\n# x/r: 0.002: 0.56334\n# x/r: 0.0025: 0.563989\n# x/r: 0.003: 0.56447\n# x/r: 0.01: 0.569822\n# x/r: 0.015: 0.56940\n# x/r: 0.012: 0.5719\n# x/r: 0.01201: 0.57192\n# 1.4499999999999993e-05 * rt**2: 0.5720702851970194\n# 0.0000145\n# z3: 10 | 07m12s | 0.57208 | 0.0205 | \n# count: 19: 0.572567, 17: 0.57263\n# ctt, stt after change: 2 | 07m56s | 0.57345 | 0.0001 | (0.00010567777727496665)\n# x4: 25 | 09m42s | 0.57359 | 0.0002 | (0.000206214286412982)\n# x4: 0.000435 (0.5737387485278771) (x4 = np.sqrt(abs(x/r)))\n# w13: 00038 (ctt,stt): 0.5737528800479372\n# ensemble of 10: 0.5772859116242378\n# ensemble of Niter=247 (random shuffle+ shift): 0.5787580886742594\n# ensemble of Niter=247 (shift only): 0.5743461440542145\n# ensemble of Niter=247 (random shuffle+ shift+ eps=0.004+vlm): 0.5865991424251623\n# 14 + ensemble: (0.0007133505234834969) 0.58787\n# w14 + ensemble: 1 | 30m13s | 0.58787 | 0.0007 | (0.0007133505234834969)\n# w14: 0.00027 (0.5873896523922799)\n# test w14, raa = x*caa + y*saa(0.00072: 0.5878990304956998)\n# test w16: r0Inv1 (21 | 21m40s | 0.58735 | 0.0000 | (1.0002801729384074e-05))\n# test w16: r0Inv1 (5.5e-06: 0.5881860039044223)\n# test r0Inv1 (5.5e-06, Niter=246, 0.5867403075395137)\n# test r0Inv1 (5.5e-06, Niter=247, 0.5872846547180826)\n# Niter = 247 (0.5880986018552999):\n# X = StandardScaler().fit_transform(np.column_stack([caa, saa, z1, z2, rt/r, x/r, y/r, z3, y1, y3, \n# ctt, stt, z4, x4, y4, raa, r0Inv]))\n# cx = [w1,w1,w2,w3, w4, w5, w6, w7, w8, w9, w10, w10, w11, w12, w13, w14, w15]\n# w15: 5.0611615056082495e-05 (17 | 21m37s | 0.58790 | 0.0001 | )\n# w15 test (5.5e-05: 0.5881768870518835)\n# w15 alone: 5.5e-05: 0.5870504337495849\n# w15 again: 5.5e-05 0.5864220587506578 (strange)\n# w15 again: 5.5e-05 (0.5880689577051738)\n# w16: 5.5e-06: 0.587602145623185 (bad since w16 was not being used)\n# after reset: w16 not being used - 0.5880689577051738\n# a2: 0.0206: 0.58157\n# org (no shift + ensemble): 0.5859135274547416\n# org (with shift + ensemble + ia1): 0.5901965251117371\n# org (with shift + ensemble + no ia1): 0.5901656684266057\n# r0Inv_d1: 7.401866174854672e-05, 0.58592 (7.5e-05: 0.5892)\n# multiprocessing - 0.6377253549867099 ( 25 mins: i7)\n# multiprocessing - z_shift (10 linspace + 1)- 0.637725354987 (32 mins)\n# z_shift (20 linspace + 1) - 0.639556619038 (44 mins)\n# z_shift ( 5 linspace + 1) - (0.62971260568) (19 mins)\n# theta0 - w15= 1.0, 1 hr 40 mins, 0.397164972826)\n# theta:0 , r0Inv using r, w15: (0.1: 0.263406003077, 0.01: 0.555401034484, 0.001:0.58657067095, 0.0001: 0.585995954153, \n# 1e-05: 0.585960618134 )\n# w15: 0.0009: 0.586267891213, 0.015: 0.586183567052, 0.002: 0.585828179853)\n# theta:0 , r0Inv using rt, w15: (0.1: 0.145904897094, 0.01: 0.536585229252, 0.001:0.585387197245, 0.0001: 0.585950889554, \n# 1e-05 to 7e-05: 0.585960618134, 40 min, 8e-05, 9e-05: 0.585950889554 )\n# theta: 0, r0Inv using r, 0.00109:0.58714214929\n# Niter : 10 , 1 min\n# Niter : 50 , 4 min\n# Niter : 75 , 5 min\n# Niter : 100 , 7 min\n# Niter : 125 , 8 min\n# Niter : 150 , 9 min (0.567703152923)\n# Niter : 200 , 11 min (0.58431499816)\n# z = 10 items, theta0, 10 items: 2h 3min 2s, 0.625977081867\n# r0Inv with rt, a1: w15 = 0.0001: 0.585950889554\n# rt/z: w15 = 1e-05: 0.585902687253\n# rt * ln(|z|): w15, 1e-05 (same for 9e-06 and 2e-05): 0.585960618134\n# ln |z|: w15: 1e-05: 0.585960618134\n# fundu: w15: 0.001: 0.58599381397\n# fundu2: same as above: w15: 0.001: 0.58599381397\n# mom: 0.01:0.585854225793, 0.001, 0.0001, 0.00001: 0.585960618134\n# mom2: 0.01: 0.585960872497, 0.001 onwards: 0.585960618134, 0.011: 0.585972357309\n# mom2 (round2): 0.001 onwards decreasing: 0.585960618134\n# mom2 trunc 4: 0.01: 0.585988797098",
"_____no_output_____"
],
[
"cluster_runs = np.random.randint(0, 50, (50, 15000))",
"_____no_output_____"
],
[
"cluster_runs.shape",
"_____no_output_____"
],
[
"consensus_clustering_labels = CE.cluster_ensembles(cluster_runs, verbose = True, N_clusters_max = 50)",
"\nINFO: Cluster_Ensembles: cluster_ensembles: due to a rather large number of cells in your data-set, using only 'HyperGraph Partitioning Algorithm' (HGPA) and 'Meta-CLustering Algorithm' (MCLA) as ensemble consensus functions.\n\n\n*****\nINFO: Cluster_Ensembles: HGPA: consensus clustering using HGPA.\n\n#\nINFO: Cluster_Ensembles: wgraph: writing wgraph_HGPA.\nINFO: Cluster_Ensembles: wgraph: 15000 vertices and 2500 non-zero hyper-edges.\n#\n\n#\nINFO: Cluster_Ensembles: sgraph: calling shmetis for hypergraph partitioning.\nINFO: Cluster_Ensembles: sgraph: (hyper)-graph partitioning completed; loading wgraph_HGPA.part.50\n#\n\nINFO: Cluster_Ensembles: cluster_ensembles: HGPA at 0.02434736018512536.\n*****\n\n*****\nINFO: Cluster_Ensembles: MCLA: consensus clustering using MCLA.\nINFO: Cluster_Ensembles: MCLA: preparing graph for meta-clustering.\nINFO: Cluster_Ensembles: MCLA: done filling hypergraph adjacency matrix. Starting computation of Jaccard similarity matrix.\nINFO: Cluster_Ensembles: MCLA: starting computation of Jaccard similarity matrix.\nINFO: Cluster_Ensembles: MCLA: done computing the matrix of pairwise Jaccard similarity scores.\n\n#\nINFO: Cluster_Ensembles: wgraph: writing wgraph_MCLA.\n#\n\n#\nINFO: Cluster_Ensembles: sgraph: calling gpmetis for graph partitioning.\nINFO: Cluster_Ensembles: sgraph: (hyper)-graph partitioning completed; loading wgraph_MCLA.part.50\n#\nINFO: Cluster_Ensembles: MCLA: delivering 50 clusters.\nINFO: Cluster_Ensembles: MCLA: average posterior probability is 0.0033593082240468202\n\nINFO: Cluster_Ensembles: cluster_ensembles: MCLA at 0.036501449656072035.\n*****\n"
],
[
"cluster_runs.shape",
"_____no_output_____"
],
[
"consensus_clustering_labels.shape",
"_____no_output_____"
],
[
"w1 = 1.1932215111905984\nw2 = 0.39740553885387364\nw3 = 0.3512647720585538\nw4 = 0.1470\nw5 = 0.01201\nw6 = 0.0003864\nw7 = 0.0205\nw8 = 0.0049\nw9 = 0.00121\nw10 = 1.4930496676654575e-05\nw11 = 0.0318\nw12 = 0.000435\nw13 = 0.00038\nw14 = 0.00072\nw15 = 5.5e-05\n# w15 = 0.000265\nw16 = 0.0031\nw17 = 0.00021\nw18 = 7.5e-05\n\nNiter=247 ",
"_____no_output_____"
],
[
"def run_make_submission():\n\n data_dir = '../data/test'\n\n\n tic = t = time.time()\n event_ids = [ '%09d'%i for i in range(0,125) ] #(0,125)\n\n if 1:\n submissions = []\n for i,event_id in enumerate(event_ids):\n hits = pd.read_csv(data_dir + '/event%s-hits.csv'%event_id)\n cells = pd.read_csv(data_dir + '/event%s-cells.csv'%event_id)\n \n labels = Fun4BO2(hits)\n\n toc = time.time()\n print('\\revent_id : %s , %0.0f min'%(event_id, (toc-tic)/60))\n\n # Prepare submission for an event\n submission = pd.DataFrame(columns=['event_id', 'hit_id', 'track_id'],\n data=np.column_stack(([event_id,]*len(hits), hits.hit_id.values, labels))\n ).astype(int)\n submissions.append(submission)\n \n for i in range(8):\n submission = extend(submission,hits)\n \n \n submission.to_csv('../cache/sub2/%s.csv.gz'%event_id,\n index=False, compression='gzip')\n\n #------------------------------------------------------\n if 1:\n\n event_ids = [ '%09d'%i for i in range(0,125) ] #(0,125)\n submissions = []\n for i,event_id in enumerate(event_ids):\n submission = pd.read_csv('../cache/sub2/%s.csv.gz'%event_id, compression='gzip')\n submissions.append(submission)\n\n \n # Create submission file\n submission = pd.concat(submissions, axis=0)\n submission.to_csv('../submissions/sub2/submission-0029.csv.gz',\n index=False, compression='gzip')\n print(len(submission))",
"_____no_output_____"
],
[
"run_make_submission()",
"event_id : 000000000 , 22 min\nevent_id : 000000001 , 48 min\nevent_id : 000000002 , 70 min\n 179.0000000"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b606c77c8b50557f7ac3685ff811941900f65b | 285,885 | ipynb | Jupyter Notebook | Bimbo/.ipynb_checkpoints/GBRT-w89-checkpoint.ipynb | zonemercy/Kaggle | 35ecb08272b6491f5e6756c97c7dec9c46a13a43 | [
"MIT"
] | 17 | 2017-10-01T00:10:19.000Z | 2022-02-07T12:11:01.000Z | Bimbo/.ipynb_checkpoints/GBRT-w89-checkpoint.ipynb | zonemercy/Kaggle | 35ecb08272b6491f5e6756c97c7dec9c46a13a43 | [
"MIT"
] | null | null | null | Bimbo/.ipynb_checkpoints/GBRT-w89-checkpoint.ipynb | zonemercy/Kaggle | 35ecb08272b6491f5e6756c97c7dec9c46a13a43 | [
"MIT"
] | 1 | 2019-08-15T03:58:51.000Z | 2019-08-15T03:58:51.000Z | 36.888387 | 30,156 | 0.487028 | [
[
[
"import pandas as pd\nimport numpy as np\nimport os\nimport math\nimport graphlab\nimport graphlab as gl\nimport graphlab.aggregate as agg\nfrom graphlab import SArray",
"_____no_output_____"
],
[
"'''钢炮'''\npath = '/home/zongyi/bimbo_data/'",
"_____no_output_____"
],
[
"train = gl.SFrame.read_csv(path + 'train_lag5.csv', verbose=False)",
"This non-commercial license of GraphLab Create for academic use is assigned to [email protected] and will expire on July 13, 2017.\n"
],
[
"town = gl.SFrame.read_csv(path + 'towns.csv', verbose=False)\ntrain = train.join(town, on=['Agencia_ID','Producto_ID'], how='left')\ntrain = train.fillna('t_c',1)\ntrain = train.fillna('tcc',0)\ntrain = train.fillna('tp_sum',0)\ndel train['Town']",
"_____no_output_____"
],
[
"del train['id']\ndel train['Venta_uni_hoy']\ndel train['Venta_hoy']\ndel train['Dev_uni_proxima']\ndel train['Dev_proxima']\ndel train['Demanda_uni_equil']\n",
"_____no_output_____"
],
[
"# relag_train = gl.SFrame.read_csv(path + 're_lag_train.csv', verbose=False)\n# train = train.join(relag_train, on=['Cliente_ID','Producto_ID','Semana'], how='left')\n# train = train.fillna('re_lag1',0)\n# train = train.fillna('re_lag2',0)\n# train = train.fillna('re_lag3',0)\n# train = train.fillna('re_lag4',0)\n# train = train.fillna('re_lag5',0)\n# del relag_train\n",
"_____no_output_____"
],
[
"# pd = gl.SFrame.read_csv(path + 'products.csv', verbose=False)\n# train = train.join(pd, on=['Producto_ID'], how='left')\n# train = train.fillna('prom',0)\n# train = train.fillna('weight',0)\n# train = train.fillna('pieces',1)\n# train = train.fillna('w_per_piece',0)\n# train = train.fillna('healthy',0)\n# train = train.fillna('drink',0)\n# del train['brand']\n# del train['NombreProducto']\n# del pd",
"_____no_output_____"
],
[
"# client = gl.SFrame.read_csv(path + 'clients.csv', verbose=False)\n# train = train.join(client, on=['Cliente_ID'], how='left')\n# del client",
"_____no_output_____"
],
[
"# cluster = gl.SFrame.read_csv(path + 'prod_cluster.csv', verbose=False)\n# cluster = cluster[['Producto_ID','cluster']]\n# train = train.join(cluster, on=['Producto_ID'], how='left')",
"_____no_output_____"
],
[
"train",
"_____no_output_____"
],
[
"# Make a train-test split\ntrain_data, test_data = train.random_split(0.999)\n\n# Create a model.\nmodel = gl.boosted_trees_regression.create(train_data, target='Demada_log',\n step_size=0.1,\n max_iterations=500,\n max_depth = 10,\n metric='rmse',\n random_seed=395,\n column_subsample=0.7,\n row_subsample=0.85,\n validation_set=test_data,\n model_checkpoint_path=path,\n model_checkpoint_interval=500)\n\n\n",
"_____no_output_____"
],
[
"model1 = gl.boosted_trees_regression.create(train, target='Demada_log',\n step_size=0.1,\n max_iterations=4,\n max_depth = 10,\n metric='rmse',\n random_seed=395,\n column_subsample=0.7,\n row_subsample=0.85,\n validation_set=None,\n resume_from_checkpoint=path+'model_checkpoint_4',\n model_checkpoint_path=path,\n model_checkpoint_interval=2)",
"_____no_output_____"
],
[
"model",
"_____no_output_____"
],
[
"w = model.get_feature_importance()",
"_____no_output_____"
],
[
"w = w.add_row_number()",
"_____no_output_____"
],
[
"w",
"_____no_output_____"
],
[
"from IPython.core.pylabtools import figsize\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport seaborn as sns\nsns.set_style('darkgrid', {'grid.color': '.8','grid.linestyle': u'--'}) \n%matplotlib inline\n\nfigsize(12, 6)\nplt.bar(w['id'], w['count'], tick_label=w['name'])\n\nplt.xticks(rotation=45)\n",
"_____no_output_____"
],
[
"# Save predictions to an SArray\npredictions = model.predict(train)\n\n# Evaluate the model and save the results into a dictionary\nresults = model.evaluate(train)\nprint results\n",
"{'max_error': 6.300516724586487, 'rmse': 0.4389403189567331}\n"
],
[
"model.summary()",
"Class : BoostedTreesRegression\n\nSchema\n------\nNumber of examples : 17797989\nNumber of feature columns : 21\nNumber of unpacked features : 21\n\nSettings\n--------\nNumber of trees : 200\nMax tree depth : 10\nTraining time (sec) : 11956.8374\nTraining rmse : 0.4465\nValidation rmse : 0.4507\n\n"
],
[
"test = gl.SFrame.read_csv(path + 'test_lag5.csv', verbose=False)\ntest = test.join(town, on=['Agencia_ID','Producto_ID'], how='left')\ndel test['Town']\ntest = test.fillna('t_c',1)\ntest = test.fillna('tcc',0)\ntest = test.fillna('tp_sum',0)",
"_____no_output_____"
],
[
"test",
"_____no_output_____"
],
[
"ids = test['id']",
"_____no_output_____"
],
[
"del test['id']",
"_____no_output_____"
],
[
"demand_log = model.predict(test)",
"_____no_output_____"
],
[
"sub = gl.SFrame({'id':ids,'Demanda_uni_equil':demand_log})",
"_____no_output_____"
],
[
"import math\nsub['Demanda_uni_equil'] = sub['Demanda_uni_equil'].apply(lambda x: math.expm1(max(0, x)))",
"_____no_output_____"
],
[
"sub",
"_____no_output_____"
],
[
"sub.save(path+'gbrt_sub3.csv',format='csv')",
"_____no_output_____"
],
[
"math.expm1(math.log1p(2))",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b607801abfa6027fe8f6527066cc342cdcf3a6 | 4,956 | ipynb | Jupyter Notebook | docs_src/utils.collect_env.ipynb | oegedijk/fastai | bd9bc96e05d464a0a4ed8719947fe941d7875e4c | [
"Apache-2.0"
] | null | null | null | docs_src/utils.collect_env.ipynb | oegedijk/fastai | bd9bc96e05d464a0a4ed8719947fe941d7875e4c | [
"Apache-2.0"
] | null | null | null | docs_src/utils.collect_env.ipynb | oegedijk/fastai | bd9bc96e05d464a0a4ed8719947fe941d7875e4c | [
"Apache-2.0"
] | null | null | null | 30.592593 | 636 | 0.577885 | [
[
[
"## Environment Checker Utils",
"_____no_output_____"
],
[
"Utilities for collecting/checking `fastai` user environment",
"_____no_output_____"
]
],
[
[
"from fastai.utils import *",
"_____no_output_____"
],
[
"from fastai.gen_doc.nbdoc import *\nfrom fastai.utils import * ",
"_____no_output_____"
],
[
"show_doc(show_install)",
"_____no_output_____"
]
],
[
[
"without nvidia-smi printout:\n\n`python -m fastai.utils.show_install`\n\nwith nvidia-smi printout:\n\n`python -m fastai.utils.show_install 1`",
"_____no_output_____"
],
[
"[`show_install`](/utils.collect_env.html#show_install)",
"_____no_output_____"
]
],
[
[
"show_doc(check_perf)",
"_____no_output_____"
]
],
[
[
"`python -m fastai.utils.check_perf`",
"_____no_output_____"
],
[
"[`check_perf`](/utils.collect_env.html#check_perf)",
"_____no_output_____"
],
[
"## Undocumented Methods - Methods moved below this line will intentionally be hidden",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
d0b60ef769f12f4ffbb0784dc2f554694745105b | 601,758 | ipynb | Jupyter Notebook | courses/dl1/lesson2-image_models.ipynb | wpride/fastai | b2a3e986de11b5a422a6efd2f48772d50d599773 | [
"Apache-2.0"
] | 1 | 2018-04-13T03:46:53.000Z | 2018-04-13T03:46:53.000Z | courses/dl1/lesson2-image_models.ipynb | wpride/fastai | b2a3e986de11b5a422a6efd2f48772d50d599773 | [
"Apache-2.0"
] | null | null | null | courses/dl1/lesson2-image_models.ipynb | wpride/fastai | b2a3e986de11b5a422a6efd2f48772d50d599773 | [
"Apache-2.0"
] | null | null | null | 712.139645 | 260,798 | 0.939157 | [
[
[
"## Multi-label classification",
"_____no_output_____"
]
],
[
[
"%reload_ext autoreload\n%autoreload 2\n%matplotlib inline",
"_____no_output_____"
],
[
"from fastai.conv_learner import *",
"_____no_output_____"
],
[
"PATH = 'data/planet/'",
"_____no_output_____"
],
[
"# Data preparation steps if you are using Crestle:\n\nos.makedirs('data/planet/models', exist_ok=True)\nos.makedirs('/cache/planet/tmp', exist_ok=True)\n\n!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train-jpg {PATH}\n!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/test-jpg {PATH}\n!ln -s /datasets/kaggle/planet-understanding-the-amazon-from-space/train_v2.csv {PATH}\n!ln -s /cache/planet/tmp {PATH}",
"_____no_output_____"
],
[
"ls {PATH}",
"\u001b[0m\u001b[01;34mmodels\u001b[0m/ \u001b[01;34mtest-jpg\u001b[0m/ \u001b[01;34mtmp\u001b[0m/ \u001b[01;34mtrain-jpg\u001b[0m/ \u001b[01;32mtrain_v2.csv\u001b[0m*\r\n"
]
],
[
[
"## Multi-label versus single-label classification",
"_____no_output_____"
]
],
[
[
"from fastai.plots import *",
"_____no_output_____"
],
[
"def get_1st(path): return glob(f'{path}/*.*')[0]",
"_____no_output_____"
],
[
"dc_path = \"data/dogscats/valid/\"\nlist_paths = [get_1st(f\"{dc_path}cats\"), get_1st(f\"{dc_path}dogs\")]\nplots_from_files(list_paths, titles=[\"cat\", \"dog\"], maintitle=\"Single-label classification\")",
"_____no_output_____"
]
],
[
[
"In single-label classification each sample belongs to one class. In the previous example, each image is either a *dog* or a *cat*.",
"_____no_output_____"
]
],
[
[
"list_paths = [f\"{PATH}train-jpg/train_0.jpg\", f\"{PATH}train-jpg/train_1.jpg\"]\ntitles=[\"haze primary\", \"agriculture clear primary water\"]\nplots_from_files(list_paths, titles=titles, maintitle=\"Multi-label classification\")",
"_____no_output_____"
]
],
[
[
"In multi-label classification each sample can belong to one or more clases. In the previous example, the first images belongs to two clases: *haze* and *primary*. The second image belongs to four clases: *agriculture*, *clear*, *primary* and *water*.",
"_____no_output_____"
],
[
"## Multi-label models for Planet dataset",
"_____no_output_____"
]
],
[
[
"from planet import f2\n\nmetrics=[f2]\nf_model = resnet34",
"_____no_output_____"
],
[
"label_csv = f'{PATH}train_v2.csv'\nn = len(list(open(label_csv)))-1\nval_idxs = get_cv_idxs(n)",
"_____no_output_____"
]
],
[
[
"We use a different set of data augmentations for this dataset - we also allow vertical flips, since we don't expect vertical orientation of satellite images to change our classifications.",
"_____no_output_____"
]
],
[
[
"def get_data(sz):\n tfms = tfms_from_model(f_model, sz, aug_tfms=transforms_top_down, max_zoom=1.05)\n return ImageClassifierData.from_csv(PATH, 'train-jpg', label_csv, tfms=tfms,\n suffix='.jpg', val_idxs=val_idxs, test_name='test-jpg')",
"_____no_output_____"
],
[
"data = get_data(256)",
"_____no_output_____"
],
[
"x,y = next(iter(data.val_dl))",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
],
[
"list(zip(data.classes, y[0]))",
"_____no_output_____"
],
[
"plt.imshow(data.val_ds.denorm(to_np(x))[0]*1.4);",
"_____no_output_____"
],
[
"sz=64",
"_____no_output_____"
],
[
"data = get_data(sz)",
"_____no_output_____"
],
[
"data = data.resize(int(sz*1.3), 'tmp')",
"_____no_output_____"
],
[
"learn = ConvLearner.pretrained(f_model, data, metrics=metrics)",
"_____no_output_____"
],
[
"lrf=learn.lr_find()\nlearn.sched.plot()",
"_____no_output_____"
],
[
"lr = 0.05",
"_____no_output_____"
],
[
"learn.fit(lr, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"lrs = np.array([lr/9,lr/3,lr])",
"_____no_output_____"
],
[
"learn.unfreeze()\nlearn.fit(lrs, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"learn.save(f'{sz}')",
"_____no_output_____"
],
[
"learn.sched.plot_loss()",
"_____no_output_____"
],
[
"sz=128",
"_____no_output_____"
],
[
"learn.set_data(get_data(sz))\nlearn.freeze()\nlearn.fit(lr, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"learn.unfreeze()\nlearn.fit(lrs, 3, cycle_len=1, cycle_mult=2)\nlearn.save(f'{sz}')",
"_____no_output_____"
],
[
"sz=256",
"_____no_output_____"
],
[
"learn.set_data(get_data(sz))\nlearn.freeze()\nlearn.fit(lr, 3, cycle_len=1, cycle_mult=2)",
"_____no_output_____"
],
[
"learn.unfreeze()\nlearn.fit(lrs, 3, cycle_len=1, cycle_mult=2)\nlearn.save(f'{sz}')",
"_____no_output_____"
],
[
"multi_preds, y = learn.TTA()\npreds = np.mean(multi_preds, 0)",
"_____no_output_____"
],
[
"files = !ls {PATH}test/",
"_____no_output_____"
]
],
[
[
"### End",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0b61a705b67ca15d5264f9fae7d7a41a37f7162 | 46,639 | ipynb | Jupyter Notebook | model/Shakespeare poetry.ipynb | aadimangla/Poetry-Generator | 5bf9db6e1bed90aacba3193cd8968f91c418bfb8 | [
"MIT"
] | 1 | 2020-09-08T07:53:04.000Z | 2020-09-08T07:53:04.000Z | model/Shakespeare poetry.ipynb | aadimangla/Poetry-Generator | 5bf9db6e1bed90aacba3193cd8968f91c418bfb8 | [
"MIT"
] | null | null | null | model/Shakespeare poetry.ipynb | aadimangla/Poetry-Generator | 5bf9db6e1bed90aacba3193cd8968f91c418bfb8 | [
"MIT"
] | null | null | null | 92.906375 | 12,472 | 0.735136 | [
[
[
"from tensorflow.keras.preprocessing.sequence import pad_sequences\nfrom tensorflow.keras.layers import Embedding, LSTM, Dense, Dropout, Bidirectional\nfrom tensorflow.keras.preprocessing.text import Tokenizer\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.optimizers import Adam\nfrom tensorflow.keras import regularizers\nimport tensorflow.keras.utils as ku \nimport numpy as np ",
"_____no_output_____"
],
[
"tokenizer = Tokenizer()\n!wget --no-check-certificate \\\n https://storage.googleapis.com/laurencemoroney-blog.appspot.com/sonnets.txt \\\n -O /tmp/sonnets.txt\ndata = open('/tmp/sonnets.txt').read()\n\ncorpus = data.lower().split(\"\\n\")\n\n\ntokenizer.fit_on_texts(corpus)\ntotal_words = len(tokenizer.word_index) + 1\n\n# create input sequences using list of tokens\ninput_sequences = []\nfor line in corpus:\n\ttoken_list = tokenizer.texts_to_sequences([line])[0]\n\tfor i in range(1, len(token_list)):\n\t\tn_gram_sequence = token_list[:i+1]\n\t\tinput_sequences.append(n_gram_sequence)\n\n\n# pad sequences \nmax_sequence_len = max([len(x) for x in input_sequences])\ninput_sequences = np.array(pad_sequences(input_sequences, maxlen=max_sequence_len, padding='pre'))\n\n# create predictors and label\npredictors, label = input_sequences[:,:-1],input_sequences[:,-1]\n\nlabel = ku.to_categorical(label, num_classes=total_words)",
"--2020-04-13 15:23:34-- https://storage.googleapis.com/laurencemoroney-blog.appspot.com/sonnets.txt\nResolving storage.googleapis.com (storage.googleapis.com)... 74.125.204.128, 2404:6800:4008:c07::80\nConnecting to storage.googleapis.com (storage.googleapis.com)|74.125.204.128|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 93578 (91K) [text/plain]\nSaving to: ‘/tmp/sonnets.txt’\n\n\r/tmp/sonnets.txt 0%[ ] 0 --.-KB/s \r/tmp/sonnets.txt 100%[===================>] 91.38K --.-KB/s in 0.001s \n\n2020-04-13 15:23:34 (130 MB/s) - ‘/tmp/sonnets.txt’ saved [93578/93578]\n\n"
],
[
"model = Sequential()\nmodel.add(Embedding(total_words, 100, input_length=max_sequence_len-1))\nmodel.add(Bidirectional(LSTM(150, return_sequences = True)))\nmodel.add(Dropout(0.2))\nmodel.add(LSTM(100))\nmodel.add(Dense(total_words/2, activation='relu', kernel_regularizer=regularizers.l2(0.01)))\nmodel.add(Dense(total_words, activation='softmax'))\nmodel.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\nprint(model.summary())\n",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, 10, 100) 321100 \n_________________________________________________________________\nbidirectional (Bidirectional (None, 10, 300) 301200 \n_________________________________________________________________\ndropout (Dropout) (None, 10, 300) 0 \n_________________________________________________________________\nlstm_1 (LSTM) (None, 100) 160400 \n_________________________________________________________________\ndense (Dense) (None, 1605) 162105 \n_________________________________________________________________\ndense_1 (Dense) (None, 3211) 5156866 \n=================================================================\nTotal params: 6,101,671\nTrainable params: 6,101,671\nNon-trainable params: 0\n_________________________________________________________________\nNone\n"
],
[
" history = model.fit(predictors, label, epochs=100, verbose=1)",
"Epoch 1/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.9055 - accuracy: 0.0205\nEpoch 2/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.4983 - accuracy: 0.0239\nEpoch 3/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.3942 - accuracy: 0.0237\nEpoch 4/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.2721 - accuracy: 0.0299\nEpoch 5/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.1864 - accuracy: 0.0362\nEpoch 6/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.1121 - accuracy: 0.0400\nEpoch 7/100\n484/484 [==============================] - 5s 10ms/step - loss: 6.0314 - accuracy: 0.0419\nEpoch 8/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.9536 - accuracy: 0.0464\nEpoch 9/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.8521 - accuracy: 0.0516\nEpoch 10/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.7521 - accuracy: 0.0556\nEpoch 11/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.6563 - accuracy: 0.0619\nEpoch 12/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.5634 - accuracy: 0.0666\nEpoch 13/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.4723 - accuracy: 0.0705\nEpoch 14/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.3826 - accuracy: 0.0779\nEpoch 15/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.2742 - accuracy: 0.0844\nEpoch 16/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.1768 - accuracy: 0.0915\nEpoch 17/100\n484/484 [==============================] - 5s 10ms/step - loss: 5.0781 - accuracy: 0.0979\nEpoch 18/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.9740 - accuracy: 0.1101\nEpoch 19/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.8732 - accuracy: 0.1164\nEpoch 20/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.7747 - accuracy: 0.1261\nEpoch 21/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.6717 - accuracy: 0.1364\nEpoch 22/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.5671 - accuracy: 0.1481\nEpoch 23/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.4530 - accuracy: 0.1610\nEpoch 24/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.3531 - accuracy: 0.1738\nEpoch 25/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.2525 - accuracy: 0.1846\nEpoch 26/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.1503 - accuracy: 0.2006\nEpoch 27/100\n484/484 [==============================] - 5s 10ms/step - loss: 4.0514 - accuracy: 0.2099\nEpoch 28/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.9520 - accuracy: 0.2227\nEpoch 29/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.8557 - accuracy: 0.2385\nEpoch 30/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.7664 - accuracy: 0.2557\nEpoch 31/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.6705 - accuracy: 0.2747\nEpoch 32/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.5791 - accuracy: 0.2914\nEpoch 33/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.4924 - accuracy: 0.3115\nEpoch 34/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.4111 - accuracy: 0.3265\nEpoch 35/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.3371 - accuracy: 0.3421\nEpoch 36/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.2465 - accuracy: 0.3648\nEpoch 37/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.1655 - accuracy: 0.3787\nEpoch 38/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.0974 - accuracy: 0.3973\nEpoch 39/100\n484/484 [==============================] - 5s 10ms/step - loss: 3.0156 - accuracy: 0.4129\nEpoch 40/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.9507 - accuracy: 0.4316\nEpoch 41/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.8838 - accuracy: 0.4453\nEpoch 42/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.8300 - accuracy: 0.4534\nEpoch 43/100\n484/484 [==============================] - 5s 11ms/step - loss: 2.7736 - accuracy: 0.4684\nEpoch 44/100\n484/484 [==============================] - 5s 11ms/step - loss: 2.7016 - accuracy: 0.4813\nEpoch 45/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.6341 - accuracy: 0.5008\nEpoch 46/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.5798 - accuracy: 0.5108\nEpoch 47/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.5262 - accuracy: 0.5217\nEpoch 48/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.4649 - accuracy: 0.5342\nEpoch 49/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.4257 - accuracy: 0.5477\nEpoch 50/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.3787 - accuracy: 0.5583\nEpoch 51/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.3209 - accuracy: 0.5676\nEpoch 52/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.2643 - accuracy: 0.5819\nEpoch 53/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.2300 - accuracy: 0.5876\nEpoch 54/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.1882 - accuracy: 0.5999\nEpoch 55/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.1475 - accuracy: 0.6087\nEpoch 56/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.0910 - accuracy: 0.6178\nEpoch 57/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.0709 - accuracy: 0.6218\nEpoch 58/100\n484/484 [==============================] - 5s 10ms/step - loss: 2.0273 - accuracy: 0.6325\nEpoch 59/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.9891 - accuracy: 0.6409\nEpoch 60/100\n484/484 [==============================] - 5s 11ms/step - loss: 1.9540 - accuracy: 0.6475\nEpoch 61/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.9178 - accuracy: 0.6557\nEpoch 62/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.8748 - accuracy: 0.6636\nEpoch 63/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.8422 - accuracy: 0.6718\nEpoch 64/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.8142 - accuracy: 0.6787\nEpoch 65/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.7803 - accuracy: 0.6868\nEpoch 66/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.7612 - accuracy: 0.6867\nEpoch 67/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.7415 - accuracy: 0.6885\nEpoch 68/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.7242 - accuracy: 0.6894\nEpoch 69/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.6808 - accuracy: 0.7058\nEpoch 70/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.6455 - accuracy: 0.7121\nEpoch 71/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.6225 - accuracy: 0.7171\nEpoch 72/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.5956 - accuracy: 0.7187\nEpoch 73/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.5835 - accuracy: 0.7218\nEpoch 74/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.5515 - accuracy: 0.7300\nEpoch 75/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.5418 - accuracy: 0.7314\nEpoch 76/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.5268 - accuracy: 0.7333\nEpoch 77/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.4872 - accuracy: 0.7451\nEpoch 78/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.4730 - accuracy: 0.7437\nEpoch 79/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.4612 - accuracy: 0.7471\nEpoch 80/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.4433 - accuracy: 0.7480\nEpoch 81/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.4318 - accuracy: 0.7509\nEpoch 82/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.4003 - accuracy: 0.7601\nEpoch 83/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3878 - accuracy: 0.7601\nEpoch 84/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3600 - accuracy: 0.7650\nEpoch 85/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3509 - accuracy: 0.7687\nEpoch 86/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3280 - accuracy: 0.7713\nEpoch 87/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3154 - accuracy: 0.7728\nEpoch 88/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3071 - accuracy: 0.7745\nEpoch 89/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.3020 - accuracy: 0.7753\nEpoch 90/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2764 - accuracy: 0.7810\nEpoch 91/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2709 - accuracy: 0.7807\nEpoch 92/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2587 - accuracy: 0.7838\nEpoch 93/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2549 - accuracy: 0.7820\nEpoch 94/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2288 - accuracy: 0.7884\nEpoch 95/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2162 - accuracy: 0.7912\nEpoch 96/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2033 - accuracy: 0.7923\nEpoch 97/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.2025 - accuracy: 0.7929\nEpoch 98/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.1857 - accuracy: 0.7945\nEpoch 99/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.1675 - accuracy: 0.7975\nEpoch 100/100\n484/484 [==============================] - 5s 10ms/step - loss: 1.1619 - accuracy: 0.8020\n"
],
[
"import matplotlib.pyplot as plt\nacc = history.history['accuracy']\nloss = history.history['loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'b', label='Training accuracy')\nplt.title('Training accuracy')\n\nplt.figure()\n\nplt.plot(epochs, loss, 'b', label='Training Loss')\nplt.title('Training loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
],
[
"seed_text = \"I Love you\"\nnext_words = 100\n \nfor _ in range(next_words):\n\ttoken_list = tokenizer.texts_to_sequences([seed_text])[0]\n\ttoken_list = pad_sequences([token_list], maxlen=max_sequence_len-1, padding='pre')\n\tpredicted = model.predict_classes(token_list, verbose=0)\n\toutput_word = \"\"\n\tfor word, index in tokenizer.word_index.items():\n\t\tif index == predicted:\n\t\t\toutput_word = word\n\t\t\tbreak\n\tseed_text += \" \" + output_word\nprint(seed_text)",
"I Love you never mark though i am young time chide last chide seen held held small moan forth torn forth back thence another needing woe brow go wrong spent grow leaves mad mad spies plot warm'd dwells sounds hits deem'd survey grow leaves new gone so dear praise twain twain told time new bright wrong did blot in mayst but bear art grow behind lie from me find make behind shame groan ' make that sky erred dispense did spend tuned spent might spend stand long short burn'd burn'd dun dun did make forth forth cherish mad forth mad seen seen reap\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b62171ea37571b32f50ce452e41149e1d0ec65 | 504,496 | ipynb | Jupyter Notebook | README.ipynb | agrouaze/sentinelrequest | 4e5514ee1538fcf513c3d06003bf2891ebd62fde | [
"MIT"
] | 1 | 2022-02-08T08:55:07.000Z | 2022-02-08T08:55:07.000Z | README.ipynb | agrouaze/sentinelrequest | 4e5514ee1538fcf513c3d06003bf2891ebd62fde | [
"MIT"
] | 2 | 2021-04-29T19:47:14.000Z | 2022-03-18T15:46:41.000Z | README.ipynb | agrouaze/sentinelrequest | 4e5514ee1538fcf513c3d06003bf2891ebd62fde | [
"MIT"
] | 3 | 2021-11-12T15:20:45.000Z | 2022-02-16T14:05:03.000Z | 281.841341 | 121,812 | 0.904291 | [
[
[
"# sentinelRequest\n\nsentinelRequest can be used to colocate a geodataframe (ie areas, trajectories, buoys, etc ...) with sentinel (1, but also 2 , 3 : all known by scihub)",
"_____no_output_____"
],
[
"## Install\n\n\n\n```\nconda install -c conda-forge lxml numpy geopandas shapely requests fiona matplotlib jupyter descartes\npip install --upgrade git+https://github.com/oarcher/sentinelrequest.git\n```\n\n",
"_____no_output_____"
],
[
"## CLI usage",
"_____no_output_____"
]
],
[
[
"!sentinelrequest --help",
"usage: sentinelrequest [-h] [--user USER] [--password PASSWORD] [--date DATE]\n [--wkt WKT] [--filename FILENAME] [--query QUERY]\n [--datatake DATATAKE] [--dateformat DATEFORMAT]\n [--dtime DTIME] [--cachedir CACHEDIR]\n [--cacherefreshrecent CACHEREFRESHRECENT] [--cols COLS]\n [--infile INFILE] [--infile_format INFILE_FORMAT]\n [--outfile OUTFILE] [--outfile_format OUTFILE_FORMAT]\n [--show] [-v]\n\nRequests SAFE list from scihub\n\noptional arguments:\n -h, --help show this help message and exit\n --user USER scihub login\n --password PASSWORD scihub password\n --date DATE date as string (see --dateformat, or date -d). if\n provided 2 time, first is start, last is stop\n --wkt WKT wkt representation of the region of interest\n --filename FILENAME filename, with joker. ex 'S1?_?W_GRD*'. default to S1*\n --query QUERY additionnal query. for exemple\n 'orbitdirection:ASCENDING AND polarisationmode:\"VV\n VH\"'\n --datatake DATATAKE retrieve adjacents datatake (ie adjacent SAFEs).\n default to 0 (no datatakes)\n --dateformat DATEFORMAT\n strftime date format. default: %Y-%m-%d %H:%M\n --dtime DTIME dtime in hours, if --date has only one date. default\n to 3\n --cachedir CACHEDIR cache dir to speedup requests\n --cacherefreshrecent CACHEREFRESHRECENT\n ignore cache if date is more recent than n days ago\n --cols COLS field output, comma separated. for ex\n --cols=index,filename. An unknown cols will show all\n available fields. 'index' is a special column name for\n index\n --infile INFILE infile (ie .csv, .gpkg, .shp ...)\n --infile_format INFILE_FORMAT\n infile format. default from file ext. see driver\n option in geopandas.to_file\n --outfile OUTFILE outfile (ie .csv, .gpkg, .shp ...)\n --outfile_format OUTFILE_FORMAT\n outfile format. default from file ext. see driver\n option in geopandas.to_file\n --show show map with matplotlib\n -v, --verbose increase output verbosity\n"
]
],
[
[
"### \"One shot\" from command line:\n\n`\n% sentinelrequest --user=xxxx --password=xxxxx --date='2018-09-23 00:00' --date='2018-09-23 12:00' --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --wkt='POLYGON ((-10 75, -10 86, 12 86, 12 84, -10 75))'\n`\n\n```\nINFO:sentinelRequest:from 2018-09-23 00:00:00 to 2018-09-23 12:00:00 : 11 SAFES\nINFO:sentinelRequest:Total : 11 SAFES\nfilename\nS1B_EW_GRDM_1SDH_20180923T071854_20180923T071954_012839_017B47_17F2.SAFE\nS1B_EW_GRDM_1SDH_20180923T071954_20180923T072054_012839_017B47_1E6F.SAFE\nS1B_EW_GRDM_1SDH_20180923T072054_20180923T072154_012839_017B47_CD41.SAFE\nS1B_EW_GRDM_1SDH_20180923T072154_20180923T072254_012839_017B47_3682.SAFE\nS1A_EW_GRDM_1SDH_20180923T081003_20180923T081107_023823_02997B_049A.SAFE\nS1A_EW_GRDM_1SDH_20180923T081107_20180923T081207_023823_02997B_6EA6.SAFE\nS1B_EW_GRDM_1SDH_20180923T085656_20180923T085756_012840_017B4E_B07B.SAFE\nS1B_EW_GRDM_1SDH_20180923T085756_20180923T085856_012840_017B4E_6CAD.SAFE\nS1B_EW_GRDM_1SDH_20180923T085856_20180923T085956_012840_017B4E_1CCD.SAFE\nS1B_EW_GRDM_1SDH_20180923T103504_20180923T103604_012841_017B54_DBBC.SAFE\nS1B_EW_GRDM_1SDH_20180923T103604_20180923T103704_012841_017B54_B267.SAFE\n```\n\n### From csv file\n\n`\n% cat test.csv\n`\n```\nindex;startdate;stopdate;geometry\narea1;2018-10-02 00:00;2018-10-02 21:00;POLYGON ((-12 35, -5 35, -5 45, -12 45, -12 35))\narea2;2018-10-13 06:00;2018-10-13 21:00;POLYGON ((-10 32, -3 32, -3 42, -10 42, -10 32))\narea3;2018-10-13 00:00;2018-10-13 18:00;POLYGON ((12 35, 5 35, 5 45, 12 45, 12 35))\n```\n\n`\n% sentinelRequest --user=xxxx --password=xxxx --infile=test.csv --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --cols=index,filename\n`\n\n```\nINFO:sentinelRequest:req 1/2 from 2018-10-02 00:00:00 to 2018-10-02 21:00:00 : 9/21 SAFES\nINFO:sentinelRequest:req 2/2 from 2018-10-13 00:00:00 to 2018-10-13 21:00:00 : 30/35 SAFES\nINFO:sentinelRequest:Total : 39 SAFES\nindex;filename\narea1;S1A_IW_GRDH_1SDV_20181002T061827_20181002T061852_023953_029DA0_C61E.SAFE\narea1;S1B_IW_GRDH_1SDV_20181002T181105_20181002T181130_012977_017F7D_FE88.SAFE\narea1;S1B_IW_GRDH_1SDV_20181002T181130_20181002T181155_012977_017F7D_93FF.SAFE\narea1;S1B_IW_GRDH_1SDV_20181002T181155_20181002T181222_012977_017F7D_CD9A.SAFE\narea3;S1A_IW_GRDH_1SDV_20181013T053545_20181013T053610_024113_02A2DB_D121.SAFE\narea3;S1A_IW_GRDH_1SDV_20181013T053815_20181013T053840_024113_02A2DB_7D53.SAFE\narea2;S1B_IW_GRDH_1SDV_20181013T062502_20181013T062527_013130_018428_1E77.SAFE\narea2;S1B_IW_GRDH_1SDV_20181013T062527_20181013T062552_013130_018428_82AB.SAFE\narea2;S1B_IW_GRDH_1SDV_20181013T062642_20181013T062707_013130_018428_AB0E.SAFE\narea2;S1B_IW_GRDH_1SDV_20181013T062707_20181013T062732_013130_018428_8210.SAFE\n```\n\nIf `--date` is specified 2 times with `--infile`, it will superseeds ones founds in infile :\n\n`\nsentinelRequest --user oarcher --password nliqt6u3 --infile=test.csv --date=last-monday-7days --date=now --filename='S1?_?W_GRD*.SAFE' --cachedir=/home1/scratch/oarcher/scihub_cache/ --cols=index,filename\n`\n\n",
"_____no_output_____"
],
[
"## API usage",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport geopandas as gpd\nimport datetime\nimport matplotlib.pyplot as plt\nimport shapely.wkt as wkt\n\n# get your own credential from https://scihub.copernicus.eu/dhus\nimport pickle\nuser,password = pickle.load(open(\"credential.pkl\",\"rb\"))\n\nimport sentinelrequest as sr\n# set default values, so we don't have to pass them at every requests\nsr.default_user = user\nsr.default_password = password\nsr.default_cachedir='/tmp/scihub_cache'\nsr.default_filename='S1?_?W_GRD*.SAFE'\n\n# optional : debug messages\n#import logging\n#sr.logger.setLevel(logging.DEBUG)",
"_____no_output_____"
],
[
"help(sr.scihubQuery)",
"Help on function scihubQuery in module sentinelrequest:\n\nscihubQuery(gdf=None, startdate=None, stopdate=None, date=None, dtime=None, timedelta_slice=None, filename=None, datatake=0, duplicate=False, query=None, user=None, password=None, min_sea_percent=None, fig=None, cachedir=None, cacherefreshrecent=None, progress=True, verbose=False, full_fig=False, alt_path=None, download=False)\n input:\n gdf : \n None or geodataframe with geometry and date. gdf usually contain almost these cols:\n index : an index for the row (for ex area name, buoy id, etc ...)\n beginposition : datetime object (startdate)\n endposition : datetime object (stopdate)\n geometry : shapely object (this one is optional for whole earth)\n date: \n column name if gdf, or datetime object\n dtime : \n if date is not None, dtime as timedelta object will be used to compute startdate and stopdate \n startdate : \n None or column name in gdf , or datetime object . not used if date and dtime are defined. \n Default to 'beginposition'\n stopdate : \n None or column name in gdf , or datetime object . not used if date and dtime are defined. \n Default to 'endposition'\n timedelta_slice:\n Max time slicing : Scihub request will be grouped or sliced to this. \n Default to datetime.timedelta(weeks=1).\n If None, no slicing is done.\n duplicate : \n if True, will return duplicates safes (ie same safe with different prodid). Default to False\n datatake : \n number of adjacent safes to return (ie 0 will return 1 safe, 1 return 3, 2 return 5, etc )\n query : \n aditionnal query string, for ex '(platformname:Sentinel-1 AND sensoroperationalmode:WV)' \n cachedir : \n cache requests for speed up. \n cacherefreshrecent : \n timedelta from now. if requested stopdate is recent, will refresh the cache to let scihub ingest new data.\n Default to datetime.timedelta(days=7).\n fig : \n matplotlib fig handle ( default to None : no plot)\n progress : True show progressbar\n verbose : False to silent messages\n alt_path : None, str or list of str\n search path in str or list of str to get safe path (columns 'path')\n str is a path string, with optionnal wilcards like `/home/datawork-cersat-public/cache/project/mpc-sentinel1/data/esa/sentinel-${missionid}/L${LEVEL}/${BEAM}/${MISSIONID}_${BEAM}_${PRODUCT}${RESOLUTION}_${LEVEL}${CLASS}/${year}/${doy}/${SAFE}`,\n or a simple path like '.' or '/tmp/scihub_download'\n if list, search in the list until a path is found.\n download : bool\n imply get_path. default to False. If True, download safes to `alt_path` (if alt_path is a list, the first index is used)\n download_wait : bool\n if `download`, will wait for non online safe to be ready. default to False.\n return :\n a geodataframe with safes from scihub, colocated with input gdf (ie same index)\n\n"
]
],
[
[
"### Simplest api usage\n\nJust a startdate and a stopdate are given, with no geometry",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n startdate=datetime.datetime(2018,10,2),\n stopdate=datetime.datetime(2018,10,3),\n fig=fig)",
"WARNING:sentinelRequest:Assuming UTC date on col beginposition\nWARNING:sentinelRequest:Assuming UTC date on col endposition\n"
]
],
[
[
"The result is a geodataframe with most information from scihub:",
"_____no_output_____"
]
],
[
[
"safes.iloc[0]",
"_____no_output_____"
]
],
[
[
"Most fields are converted from str to python type (geometry, datetime, int ...)",
"_____no_output_____"
]
],
[
[
"safes.iloc[1:4]['footprint'].plot()",
"_____no_output_____"
],
[
"print('safe was ingested %s after aquisition' % (safes.iloc[0]['ingestiondate']-safes.iloc[0]['endposition']))",
"safe was ingested 0 days 05:50:26.096000 after aquisition\n"
]
],
[
[
"### Using a geodataframe with geometries \n\nAs an example, two areas are defined. Note that the index is named with the area name\n",
"_____no_output_____"
]
],
[
[
"gdf = gpd.GeoDataFrame({\n \"beginposition\" : [ datetime.datetime(2018,10,2,0) , datetime.datetime(2018,10,13,0) ],\n \"endposition\" : [ datetime.datetime(2018,10,2,21) ,datetime.datetime(2018,10,13,18) ],\n \"geometry\" : [ wkt.loads(\"POINT (-7.5 53)\").buffer(4), wkt.loads(\"POLYGON ((-12 35, -5 35, -5 45, -12 45, -12 35))\")] \n },index=[\"Irland\",\"Portugal\"])\ngdf",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n gdf=gdf,\n min_sea_percent=20, \n fig=fig)\n",
"WARNING:sentinelRequest:Assuming UTC date on col beginposition\nWARNING:sentinelRequest:Assuming UTC date on col endposition\nWARNING:sentinelRequest:no crs provided. assuming lon/lat with greenwich/antimeridian handling\nINFO:sentinelRequest:Slicing into 2 chunks of 7 days, 0:00:00 ...\nINFO:sentinelRequest:Slicing done in 0.0s . 2/2 non empty slices.\n"
]
],
[
[
"User requested area are in green, and found safes are in blue.\n",
"_____no_output_____"
],
[
"Index from original request are preserved, so it's easy to know the area that belong to a safe. (See end of example 2 for advanced index handling).",
"_____no_output_____"
]
],
[
[
"safes.loc['Portugal']",
"_____no_output_____"
]
],
[
[
"### Working with projection\n\nSentinelRequest works with projections, by defining crs in gdf.\n\nThe colocalisation is done using this crs.\n\nget safes around 1000km, at 84° (North pole included) ",
"_____no_output_____"
]
],
[
[
"import pyproj\ngdf = gpd.GeoDataFrame({\n \"beginposition\" : [ datetime.datetime(2019,12,1,0) ],\n \"endposition\" : [ datetime.datetime(2019,12,4,0)],\n \"geometry\" : [ wkt.loads(\"POINT (0 84)\")] \n },index=[\"Artic\"], crs=pyproj.CRS('epsg:4326'))\n\n\n# to polar projection (units in meters)\ngdf.to_crs(pyproj.CRS('epsg:3408'), inplace=True)\ngdf.loc[\"Artic\",\"geometry\"]=gdf.loc[\"Artic\"].geometry.buffer(1000 * 1000)\n\n\n\nfig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n gdf=gdf,\n min_sea_percent=20, \n fig=fig)",
"WARNING:sentinelRequest:Assuming UTC date on col beginposition\nWARNING:sentinelRequest:Assuming UTC date on col endposition\n"
]
],
[
[
"### Cyclone track colocalization",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n#ibtracs=gpd.read_file('tmp/IBTrACS.NA.list.v04r00.points.shp')\n#gdf_track=ibtracs[ibtracs['SID'] == '2019235N10324']\n#gdf_track=gdf_track[['ISO_TIME','USA_WIND','geometry']]\n#gdf_track['ISO_TIME']=pd.to_datetime(gdf_track['ISO_TIME'],format=\"%Y-%m-%d %H:%M:%S\")\n#gdf_track.reset_index(inplace = True,drop=True) \n#gdf_track.to_file(\"track.gpkg\", driver=\"GPKG\")\n\ngdf_track = gpd.read_file('track.gpkg')\ngdf_track['ISO_TIME']=pd.to_datetime(gdf_track['ISO_TIME'],format=\"%Y-%m-%d %H:%M:%S\")\ngdf_track",
"/home/oarcher/anaconda3/envs/xsar/lib/python3.9/site-packages/geopandas/geodataframe.py:577: RuntimeWarning: Sequential read of iterator was interrupted. Resetting iterator. This can negatively impact the performance.\n for feature in features_lst:\n"
],
[
"fig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n gdf=gdf_track,\n date='ISO_TIME', # no startdate/stopdate, but a date ans a dtime\n dtime=datetime.timedelta(hours=1.5),\n datatake=1, # take adjacents safes, up to one.\n fig=fig)",
"INFO:sentinelRequest:Slicing into 3 chunks of 7 days, 0:00:00 ...\nINFO:sentinelRequest:Slicing done in 0.1s . 3/3 non empty slices.\n"
]
],
[
[
"#### datatake\n\nHere, `datatake=1` is specified to retrieve adjacents safes from colocated ones (in cyan). When specified, the result contain a `datatake_index` column. 0 means the colocated one, and other values are the range of the adjacent safe (up to -n..n with `datatake=n`)\n\nPositive `datatake_index` are for safes *after* the colocated one, and negative index are fo safes *before* the colocated one.",
"_____no_output_____"
]
],
[
[
"safes[['filename','datatake_index']]",
"_____no_output_____"
]
],
[
[
"#### Time slicing with timedelta_slice\n\nOne can see on previous figure that 3 requests are done. gdf rows are grouped to reduce the amount of scihub requests with the `timedelta_slice` parameter (default to `datetime.timedelta(weeks=1)` )\n\nIf we reduce `timedelta_slice`, we can see that more scihub request are done, with less uncolocated safes (ie yellow). (be warned with a big `timedelta_slice` : this can produce scihub timeouts).\n\n(with `timedelta_slice=None`, this feature is *disabled* : a scihub request is done for *every* geometry).",
"_____no_output_____"
]
],
[
[
"# same request as above, but with reduced timedelta_slice\nfig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n gdf=gdf_track,\n date='ISO_TIME',\n dtime=datetime.timedelta(hours=1.5),\n timedelta_slice=datetime.timedelta(days=1),\n datatake=1,\n full_fig = True, # to show internals requests and colocs\n fig=fig)",
"INFO:sentinelRequest:Slicing into 18 chunks of 1 day, 0:00:00 ...\nINFO:sentinelRequest:Slicing done in 0.1s . 18/18 non empty slices.\n"
]
],
[
[
"#### Merging source and result with shared index\n\nAs seen before, the result (safes) share the same index as the source. So we can merge the two geodataframe, to associate a wind speed from the cyclone track with the safe, and compute distance from the eye to the safe.",
"_____no_output_____"
]
],
[
[
"# here, we merge the result with the source request, to associate wind speed to each safe.\nmerged=safes[['filename','datatake_index','footprint']].merge(\n gdf_track[['USA_WIND','geometry']],left_index=True,right_index=True)\nmerged['eye_dist'] = merged.set_geometry('geometry').distance(merged.set_geometry('footprint').exterior)\n# negative dist if safe contains eye\nmerged['eye_dist']=merged['eye_dist']*(((~merged.set_geometry('footprint').contains(merged.set_geometry('geometry'))+1)*2)-3)\nmerged[['filename','datatake_index','USA_WIND','eye_dist']]\n",
"<ipython-input-16-6297d856a9af>:4: UserWarning: Geometry is in a geographic CRS. Results from 'distance' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.\n\n merged['eye_dist'] = merged.set_geometry('geometry').distance(merged.set_geometry('footprint').exterior)\n"
]
],
[
[
"## Annexes\n\n### Antimeridian handling: small geometry vs large one\n\n\nGiven 2 points on the earth, there is two possible paths: one short, and one long that wrap around the earth.\nNote: only longitude is wrapped, as if earth was a cylinder (epgs 4326 used for computation) \n\nBy default, geometry are the smallest ones. To preserve a large geometry, GeometryCollection must be used.\n",
"_____no_output_____"
]
],
[
[
"from shapely.geometry import GeometryCollection\n\n# the polygon is more than 180 deg wide. It will be wrapped, and will cross antimeridian\nlarge_poly = wkt.loads(\"POLYGON ((-140 -14, 140 -14, 140 -20, -140 -20, -140 -14))\")\n\ngdf = gpd.GeoDataFrame({\n \"beginposition\" : [ datetime.datetime(2018,10,1)],\n \"endposition\" : [ datetime.datetime(2018,10,31) ],\n \"geometry\" : [ large_poly ] \n },index=[0])\n\nfig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n gdf=gdf,\n fig=fig)\nplt.show()\n\n# same polygon, but encapsulated in a GeometryCollection : it will not be wrapped\ngdf = gpd.GeoDataFrame({\n \"beginposition\" : [ datetime.datetime(2018,10,1)],\n \"endposition\" : [ datetime.datetime(2018,10,31) ],\n \"geometry\" : [ GeometryCollection([large_poly]) ] \n },index=[0])\n\nfig = plt.figure(figsize=(10,7))\nsafes = sr.scihubQuery(\n gdf=gdf,\n fig=fig)\nplt.show()\n\ngdf",
"WARNING:sentinelRequest:Assuming UTC date on col beginposition\nWARNING:sentinelRequest:Assuming UTC date on col endposition\nWARNING:sentinelRequest:no crs provided. assuming lon/lat with greenwich/antimeridian handling\nINFO:sentinelRequest:Slicing into 5 chunks of 7 days, 0:00:00 ...\nINFO:sentinelRequest:Slicing done in 0.1s . 5/5 non empty slices.\n"
],
[
"import shapely.ops\nlen(shapely.ops.unary_union(gdf_track.geometry).buffer(2).simplify(1.9).wkt)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0b635a2ac30ee954747bd799ecdecf85f92a148 | 206,619 | ipynb | Jupyter Notebook | Autopilot/Autopilot_replicated.ipynb | Hira63S/DeepLearningResearch | b6e8298a88fbc81de06d8e202603a80af8bbdaa2 | [
"MIT"
] | null | null | null | Autopilot/Autopilot_replicated.ipynb | Hira63S/DeepLearningResearch | b6e8298a88fbc81de06d8e202603a80af8bbdaa2 | [
"MIT"
] | null | null | null | Autopilot/Autopilot_replicated.ipynb | Hira63S/DeepLearningResearch | b6e8298a88fbc81de06d8e202603a80af8bbdaa2 | [
"MIT"
] | null | null | null | 309.30988 | 145,904 | 0.909805 | [
[
[
"# Reproduct Autopilot Architecture\n\nThe Autopilot has the following Architecture:\n~ ResNet50-like backbone\n~ FPN - DeepLabV3- UNet - like heads\n~ 15 tasks\n ~ subtasks i.e. if task is car detection, then the sub task is what kind of car, is it stationary? Parked, broken down?\n\nFor later exploration:\nStitching up of images across space and time happens inside RNNs.\nAlso explore Faster R-CNNs but they have lower inference rate in real-time detection etc.\n",
"_____no_output_____"
],
[
"## First Step:\nUse transfer learning to load ResNet-50 model because training it and loading it that way has been a hassle.\n\nWe are most likely using feature extraction techniques to get the data from resnet backbone and passing them onto the tasks",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.optim as optim\nimport torchvision\nfrom torchvision import models, transforms\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"batch_size = 50\n\n# get the CIFAR-10 images:\n\ntrain_data_transform = transforms.Compose([\n transforms.Resize(224),\n transforms.RandomHorizontalFlip(),\n transforms.RandomVerticalFlip(),\n transforms.ToTensor(),\n transforms.Normalize((0.4914, 0.4821, 0.4465), (0.2470, 0.2435, 0.2616))\n])\n\ntrain_set = torchvision.datasets.CIFAR10(root='./data',\n train=True, download = True,\n transform=train_data_transform)\n\ntrain_loader = torch.utils.data.DataLoader(train_set, batch_size=batch_size,\n shuffle=True, num_workers=2)\n",
"Files already downloaded and verified\n"
]
],
[
[
"## CIFAR10 Data\n10000 x 3072 numpy arrays. i.e 1024 values i.e. 32x32 image and since there are 3 channels, we get 3072 shaped numpy arrays.\nEach row in the array stores a 32x32 colour image\nThe first 1024 entries contain the red channel values, the next 1024 are green and the next 1024 are blue.\n",
"_____no_output_____"
]
],
[
[
"def imshow(img):\n img = img/2 + 0.5 # unnormalize\n npimg = img.numpy()\n plt.imshow(np.transpose(npimg, (1,2,0)))\n plt.show()\n ",
"_____no_output_____"
],
[
"dataiter = iter(train_loader)\nimages, labels = dataiter.next()",
"_____no_output_____"
],
[
"classes = ('plane', 'car', 'bird', 'cat',\n 'deer', 'dog', 'frog', 'horse', 'ship', 'truck')",
"_____no_output_____"
],
[
"imshow(torchvision.utils.make_grid(images))\nprint(' '.join('%5s'%classes[labels[j]] for j in range(50)))",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
],
[
"imshow(images[49])",
"Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).\n"
],
[
"len(train_loader)",
"_____no_output_____"
],
[
"val_data_transform = transforms.Compose([\n transforms.Resize(224),\n transforms.ToTensor(),\n transforms.Normalize((0.4914, 0.4821, 0.4465), (0.2470, 0.2435, 0.2616))\n])\nval_set = torchvision.datasets.CIFAR10(root='./data',\n train=False, download=True,\n transform=val_data_transform)\nval_order = torch.utils.data.DataLoader(val_set,\n \n batch_size=batch_size,\n shuffle=False, num_workers=2\n )",
"Files already downloaded and verified\n"
],
[
"device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
],
[
"def train_model(model, loss_function, optimizer, data_loader):\n overall_start = timer()\n # set the model mode\n # model.train()\n for epoch in range(n_epochs):\n current_acc = 0\n current_loss =0\n model.train()\n start = timer()\n \n \n # iterate over the examples in the dataset:\n for i, (inputs, labels) in enumerate(data_loader):\n # send them to the GPU first\n inputs = inputs.to(device)\n labels = labels.to(device)\n \n # zero the parameter gradients\n optimizer.zero_grad()\n \n with torch.set_grad_enabled(True):\n # forward\n outputs = model(inputs)\n _, predictions = torch.max(outputs, 1)\n loss = loss_function(outputs, labels)\n \n # backward\n loss.backward()\n optimizer.step()\n \n # statistics\n current_loss += loss.item() * inputs.size(0)\n current_acc += torch.sum(predictions == labels.data)\n \n total_loss = current_loss / len(data_loader.dataset)\n total_acc = current_acc.double() / len(data_loader.dataset)\n \n print('Train Loss: {:.4f}; Accuracy {:.4f}'.format(total_loss, total_acc))",
"_____no_output_____"
],
[
"def test_model(model, loss_function, data_loader):\n # set model in evaluation mode\n model.eval()\n \n current_loss = 0.0\n current_acc = 0\n \n # iterate over the validation data\n for i, (inputs, labels) in enumerate(data_loader):\n inputs = inputs.to(device)\n labels = labels.to(device)\n \n with torch.set_grad_enabled(False):\n outputs = model(inputs)\n _, predictions = torch.max(outputs, 1)\n loss = loss_function(outputs, labels)\n \n # statistics\n current_loss += loss.item() * inputs.size(0)\n current_acc += torch.sum(predictions == labels.data)\n \n total_loss = current_loss / len(data_loader.dataset)\n total_acc = current_acc.double()/ len(data_loader.dataset)\n \n print('Test Loss: {:.4f}; Accuracy {:.4f}'.format(total_loss, total_acc))\n \n return total_loss, total_acc\n \n ",
"_____no_output_____"
]
],
[
[
"Now, onto the transfer learning scenario where we are going to use the pretrained network as a feature extractor.\n1. Let's use ResNet50\n2. Replace last layer of the model with a new layer with 10 outputs\n3. Exclude the existing network layers from the backward pass and only pass the newly added fully-connected layer to the Adam optimizer.\n4. Run the training for epochs and evaluate the network accuracy after each epoch.\n5. Plot the test accuracy",
"_____no_output_____"
],
[
"# Load Resnet50",
"_____no_output_____"
]
],
[
[
"model = torchvision.models.resnet50(pretrained=True)\nmodel.eval()",
"_____no_output_____"
],
[
"def train(model, criterion,\n optimizer,\n train_loader,\n valid_loader,\n save_file_name,\n max_epochs_stop=3,\n n_epochs = 20,\n print_every=2):\n \"\"\"Train the Pytorch model while including the \"\"\"",
"_____no_output_____"
],
[
"def tl_feature_extractor(epochs=5):\n # load the pretrained model\n model = torchvision.models.resnet50(pretrained=True)\n \n # exclude the existing parameters from backward pass\n # for performance\n for param in model.parameters():\n param.requires_grad = False\n \n # newly constructed layers have requires_grad=True by default\n num_features = model.fc.in_features\n model.fc = nn.Sequential(\n nn.Linear(num_features, 1000),\n nn.ReLU(),\n nn.Dropout(0.4),\n nn.Linear(1000, 10),\n nn.LogSoftmax(dim=1))\n# model.fc = nn.Linear(1000, 10)\n \n # transfer to GPU\n model = model.to(device)\n \n loss_function = nn.CrossEntropyLoss()\n \n # only parameters of the final layer are being optimized\n optimizer = optim.Adam(model.fc.parameters()) # otherwise, it would be just model.parameters()\n \n # setting timing\n overall_start = timer()\n # train\n test_acc = list()\n for epoch in range(epochs):\n print('Epoch {}/{}'.format(epoch+1, epochs))\n \n train_model(model, loss_function, optimizer, train_loader)\n _, acc = test_model(model, loss_function, val_order)\n test_acc.append(acc)\n \n plot_accuracy(test_acc)",
"_____no_output_____"
]
],
[
[
"model.classifier[6] = nn.Sequential(\n nn.Linear(n_inputs, 256), \n nn.ReLU(), \n nn.Dropout(0.4),\n nn.Linear(256, n_classes), \n nn.LogSoftmax(dim=1))",
"_____no_output_____"
]
],
[
[
"tl_feature_extractor()",
"Epoch 1/5\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"raw"
],
[
"code"
]
] |
d0b646b578fa94a0d5aea3fcd363011e42c248d4 | 296,571 | ipynb | Jupyter Notebook | BA_775_Team_Assignment_Team_4b.ipynb | Amberineee/ecommerce_covid_analysis | 3618127117bf36838dc554f1bf0a4b6bb07af2e2 | [
"MIT"
] | null | null | null | BA_775_Team_Assignment_Team_4b.ipynb | Amberineee/ecommerce_covid_analysis | 3618127117bf36838dc554f1bf0a4b6bb07af2e2 | [
"MIT"
] | null | null | null | BA_775_Team_Assignment_Team_4b.ipynb | Amberineee/ecommerce_covid_analysis | 3618127117bf36838dc554f1bf0a4b6bb07af2e2 | [
"MIT"
] | null | null | null | 103.515183 | 99,966 | 0.752383 | [
[
[
"<a href=\"https://colab.research.google.com/github/Amberineee/ecommerce_covid_analysis/blob/main/BA_775_Team_Assignment_Team_4b.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import auth\nauth.authenticate_user()",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
]
],
[
[
"#E-commerce Dataset 2020\n\n**For this project, we want to enhance the webpage with the provided dataset of e-Commerce dataset in February and April by taking in consideration COVID-19 pandemic.**\n\nGoal: Our goal is analyzing customer online purchasing behavior during pre covid-19 and post covid-19 using February and April dataset, respectively. Which allows us to understand the traffic of the website and conversion made by consumers to potential solve E-commerce’s dataset problems, and expand the opportunities for marketing campaigns, target promotions and optimizing the inventory level.\n\nDataset: https://www.kaggle.com/mkechinov/ecommerce-behavior-data-from-multi-category-store",
"_____no_output_____"
],
[
"##First, Let's have a look for the month of February (Pre-COVID):",
"_____no_output_____"
],
[
"###To begin, we will identify the Top 10 popular brands that are purchased by the consumers for February 2020.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT brand, count(brand) AS total_brand_purchase\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020` \nWHERE event_type = 'purchase'\nGROUP BY brand\nORDER BY count(brand) DESC\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Results: We noticed that Samsung is consumer’s favorite brand, followed by Apple and Xiaomi. All electronic brands!",
"_____no_output_____"
],
[
"###Next, let’s give a look to the conversion rate for the month of February from 2020.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT \nCOUNTIF(event_type = 'purchase')/COUNT(event_type) AS conversion_rate, \ncategory_code,\nROUND(sum(price), 2) AS total_Price\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020` \nGROUP BY category_code\nORDER BY Conversion_rate DESC\nlimit 10",
"_____no_output_____"
]
],
[
[
"Results: It shows that the highest conversion rate is the construction tools with a rate of 0.0397, followed by apparel with a rate of 0.0334.",
"_____no_output_____"
],
[
"###We also want to identify who are the Top consumers in the ecommerce webpage, and how much do they spend for this month, February.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT user_id, ROUND(SUM(price)) AS total_spending_FEB\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\nWHERE event_type = 'purchase'\nGROUP BY user_id\nORDER BY total_spending_FEB DESC\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Result: our Top consumer identified as user_id: '563051763' spent around $302,726.0 on the webpage.",
"_____no_output_____"
],
[
"###Now, let’s check the average of all users’ spendings for the month of February.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT ROUND(AVG(price), 2) as avg_spending_feb\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\nWHERE event_type = 'purchase'\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Result: The average of users’ spendings for the month of February is $317.57.",
"_____no_output_____"
],
[
"###After identifying the highest consumer as user_id: '563051763', we want to compare their spending with the average of all users’ spendings for the month of February.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT user_id AS USER_ID, ROUND(SUM(price)) AS Total_spending, COUNT(user_session) as Sessions, \n(\nSELECT round(AVG(price), 2) as avg_spending\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\nWHERE event_type = 'purchase'\n) as Avg_spending_of_all_users\n \n FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\n WHERE user_id = 563051763 and event_type = 'purchase'\n GROUP BY user_id",
"_____no_output_____"
]
],
[
[
"This query shows the top buyer who have spend the most in this ecommerce website, number of sessions, and the average of all users' spendings.",
"_____no_output_____"
],
[
"## Now, we would check for the month of April (post-COVID):",
"_____no_output_____"
],
[
"###For the month of April, 2020. We will identify the Top 10 popular brands that were purchased by the consumers during post COVID.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT brand, count(brand) AS total_brand_purchase\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE event_type = 'purchase'\nGROUP BY brand\nORDER BY count(brand) DESC\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Results: We noticed that the results are the same as pre-COVID (February). Samsung is still consumer’s favorite brand, followed by Apple and Xiaomi. All electronic brands!",
"_____no_output_____"
],
[
"###Let’s give a look to the conversion rate for the month of April.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT countif(event_type = 'purchase')/count(event_type) AS Conversion_rate, category_code, sum(price) Total_Price\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nGROUP BY category_code\nORDER BY Conversion_rate DESC\nlimit 10",
"_____no_output_____"
]
],
[
[
"Results: It shows that the highest conversion rate is the stationery paper with a rate of 0.038, followed by kitchen appliances with a small difference of 0.001.",
"_____no_output_____"
],
[
"###Next, we want to identify who are the Top consumers in the ecommerce webpage, and how much they spend.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT user_id, round(SUM(price), 2) as total_spending\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE event_type = 'purchase'\nGROUP BY user_id\nORDER by total_spending DESC\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Results: our Top consumer identified as user_id: '553446649' spent around $122,525.19 on the webpage, which is way more lower than in February. This leads to a better understanding of consumers' behaviour that due the pandemic consumers had shifted their values, and cutting down unneed purchases to essentials.",
"_____no_output_____"
],
[
"###Now, let’s check the average of all users’ spendings for the month of April.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT ROUND(AVG(price), 2) as avg_spending_of_all_users\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE event_type = 'purchase'",
"_____no_output_____"
]
],
[
[
"Result: As we expected, the average of users’ spendings also decreased during pandemic. Average for April is `$252.93`, while for Feb was `$317.57`.",
"_____no_output_____"
],
[
"###After identifying the highest consumer as user_id: '553446649', we want to compare their spending with the average of all users’ spendings for the month of April.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT user_id AS USER_ID, ROUND(SUM(price)) AS TOTAL_SPENDING,COUNT(user_session) AS TOTA_SESSIONS, \n(\nSELECT round(AVG(price), 2) as avg_spending\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE event_type = 'purchase'\n) as AVG_SPENDING_OF_ALL_USERS\n \n FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\n WHERE user_id = 553446649 and event_type = 'purchase'\n GROUP BY user_id",
"_____no_output_____"
]
],
[
[
"This query shows the top buyer who have spend the most in this ecommerce website, and the average of all users' spendings",
"_____no_output_____"
],
[
"##Let's check deeper the insights by combining both datasets!",
"_____no_output_____"
],
[
"###We also want to know how many times they shop on this website in February and April.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT \nuser_id,\nCOUNT(user_id) AS returning\nFROM\n(\nSELECT user_id, event_type\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020` AS FEB\nUNION ALL\nSELECT user_id, event_type\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020` AS APR\n)\nWHERE event_type = 'purchase'\nGROUP BY user_id\nHAVING COUNT(user_id) > 1\nORDER BY returning DESC\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Results: Our top consumer identified as user_id: ‘609817194’ returned back to the website 573 times. Surprisingly, our top consumer from the month of February and April are not in the Top 10.",
"_____no_output_____"
],
[
"###Now we want to see the impact of COVID-19 by finding the percentage change in sales for popular categories in February",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT \ncategory_code,\n(total_sales_apr - total_sales_feb)/total_sales_feb AS percent_change\nFROM\n(\nSELECT *\nFROM \n (SELECT \ncategory_code,\nSUM(price) AS total_sales_apr\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE category_code IS NOT NULL AND event_type = 'purchase'\nGROUP BY category_code\nORDER BY total_sales_apr DESC\n) AS apr\n \nINNER JOIN \n \n (SELECT \ncategory_code AS cat,\nSUM(price) AS total_sales_feb,\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020` \nWHERE category_code IS NOT NULL and event_type = 'purchase'\nGROUP BY category_code\nORDER BY total_sales_feb DESC) AS feb\n \nON apr.category_code = feb.cat\n)\nORDER BY total_sales_feb DESC\nLIMIT 10\n",
"_____no_output_____"
]
],
[
[
"Results: Most popular categories have lower sales in April than in February. But headphone, massager and refrigerators categories have higher sales in April. It might casued by working from home.\n",
"_____no_output_____"
],
[
"# Are views correlated with the number of speding in each month?",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nselect count(event_type) as feb_visits, \n\n(select round(sum(price),2) as profit_from_feb from `ba775-team-4b.4b_dataset.ecommerce_feb_2020` where event_type = 'purchase')feb_profit,\n\n(select count(event_type) as total_visits_april FROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere event_type = 'view'\ngroup by event_type) april_visits,\n\n(select round(sum(price),2) as profit_from_april from `ba775-team-4b.4b_dataset.ecommerce_april_2020` where event_type = 'purchase') april_profit\n\n FROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\nwhere event_type = 'view'\ngroup by event_type",
"_____no_output_____"
]
],
[
[
"There is a negative correlation between the number of views and consumer's spending on each month.\n\nResults: April visits times increased by 9.8% since February, but decreased sales by 21.84%.",
"_____no_output_____"
],
[
"###Also, we wants to investigate how well samsung's top selling categories in February performed in April",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT \nA.*,FEB_total_sales,round(APR_total_sales-FEB_total_sales,2) FEB_APR_SalesDifferences, \nround(((APR_total_sales-FEB_total_sales)/FEB_total_sales),2) FEB_APR_SalesChanges_percentage\nFROM\n(SELECT A.*,B.APR_total_sales\nFROM \n(SELECT A.brand,A.category_code,count(A.brand) AS count_APR_brand_sales, count_FEB_brand_sales\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`A\n\nLEFT JOIN \n(SELECT brand,category_code,count(brand) AS count_FEB_brand_sales\nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\nWHERE event_type = 'purchase' and brand = 'samsung' AND category_code IS NOT NULL \nGROUP BY category_code,brand\n) B\nUSING(category_code)\n\nWHERE event_type = 'purchase' and A.brand = 'samsung' AND category_code IS NOT NULL\nGROUP BY category_code,brand,count_FEB_brand_sales \nORDER BY count_APR_brand_sales DESC\n\nlimit 15)\nA\nLEFT JOIN\n(SELECT category_code,round(sum(price),2) as APR_total_sales \nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE brand = 'samsung' AND event_type = 'purchase' AND category_code IS NOT NULL\nGROUP BY category_code\n) B\nUSING(category_code)\n) A\n\nLEFT JOIN \n(\nSELECT category_code,round(sum(price),2) as FEB_total_sales \nFROM `ba775-team-4b.4b_dataset.ecommerce_feb_2020`\nWHERE brand = 'samsung' AND event_type = 'purchase' AND category_code IS NOT NULL\nGROUP BY category_code\n) B\nUSING(category_code)\n",
"_____no_output_____"
]
],
[
[
"Result: most of the February's top-selling categories for samsung increased a lot in April, especially for air conditioner category.",
"_____no_output_____"
],
[
"# Number of products they purchased in different category.\n\n\n",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT user_id,num_of_accessories,num_of_apparel,num_of_appliances,num_of_auto,num_of_computers,num_of_construction,num_of_country_yard,num_of_electronics,num_of_furniture,num_of_kids,num_of_medicine,num_of_sport,num_of_stationery from\n(SELECT user_id, count(user_id) as num_of_accessories\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"accessories%\" and event_type=\"purchase\"\ngroup by user_id) as accessories\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_apparel\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"apparel%\" and event_type=\"purchase\"\ngroup by user_id) as apparel\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_appliances\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"appliances%\" and event_type=\"purchase\"\ngroup by user_id) as appliances\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_auto\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"auto%\" and event_type=\"purchase\"\ngroup by user_id) as auto\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_computers\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"computers%\" and event_type=\"purchase\"\ngroup by user_id) as computers\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_construction\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"construction%\" and event_type=\"purchase\"\ngroup by user_id) as construction\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_country_yard\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"country_yard%\" and event_type=\"purchase\"\ngroup by user_id) as country_yard\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_electronics\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"electronics%\" and event_type=\"purchase\"\ngroup by user_id) as electronics\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_furniture\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"furniture%\" and event_type=\"purchase\"\ngroup by user_id) as furniture\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_kids\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"kids%\" and event_type=\"purchase\"\ngroup by user_id) as kids\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_medicine\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"medicine%\" and event_type=\"purchase\"\ngroup by user_id) as medicine\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_sport\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"sport%\" and event_type=\"purchase\"\ngroup by user_id) as sport\nusing(user_id)\nFULL OUTER JOIN\n(SELECT user_id, count(user_id) as num_of_stationery\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nwhere category_code like \"stationery%\" and event_type=\"purchase\"\ngroup by user_id) as stationery\nusing(user_id)\norder by num_of_computers desc",
"_____no_output_____"
]
],
[
[
"# Abandon Rate",
"_____no_output_____"
]
],
[
[
"%%bigquery --project ba775-team-4b\nSELECT a.user_id, \na.cart_count, \nifnull(b.purchase_count,0) as purchase_count, \nround((ifnull(b.purchase_count,0)/a.cart_count),3)as purchase_rate,\nround((1-(ifnull(b.purchase_count,0)/a.cart_count)),3) as abandon_rate\nFROM `ba775-team-4b.4b_dataset.cart_count_per_user_id` a \nleft join \n`ba775-team-4b.4b_dataset.purchase_count_per_user_id` b\nusing (user_id)\nwhere a.cart_count>=b.purchase_count and round((1-(ifnull(b.purchase_count,0)/a.cart_count)),3)\t> 0.9\norder by abandon_rate desc",
"_____no_output_____"
],
[
"%%bigquery --project ba775-team-4b\nSELECT a.user_id, \na.cart_count, \nifnull(b.purchase_count,0) as purchase_count, \nround((ifnull(b.purchase_count,0)/a.cart_count),3)as purchase_rate,\nround((1-(ifnull(b.purchase_count,0)/a.cart_count)),3) as abandon_rate\nFROM `ba775-team-4b.4b_dataset.purchase_per_userid _april` a \nleft join \n`ba775-team-4b.4b_dataset.purchase_count_per_userid_april` b\nusing (user_id)\nwhere a.cart_count>=b.purchase_count and round((1-(ifnull(b.purchase_count,0)/a.cart_count)),3)\t> 0.9\norder by abandon_rate desc",
"_____no_output_____"
]
],
[
[
"The abandon rate can be used as the metric to evaluate the purchase power of the users. The abandon rate of a certain user = 1-(number of 'purchase' event /number of 'cart' event). Only the user info with high abandon rate(#>0.9) is included and have the potential to work on increasing sales conversion, such as sending target promotions, poping promotional codes, running market campaigns, etc. \n\nNote: We noticed that for some users purchased more than they carted and will make abandon rate calculated less than 0, which can be high indication of purchase power. Hereby we dropped those users as they are not target audience for this part. ",
"_____no_output_____"
],
[
"# Machine Learning",
"_____no_output_____"
]
],
[
[
"# Using product features\nCREATE MODEL\n model.products\nOPTIONS\n ( model_type = 'kmeans',\n num_clusters = 2,\n distance_type = 'euclidean') AS\nSELECT \ncategory_code, brand, price, count(product_id)\nFROM `ba775-team-4b.4b_dataset.ecommerce_april_2020`\nWHERE event_type='purchase'\nGROUP BY category_code, product_id, brand, price",
"_____no_output_____"
],
[
"# Using users features\n## have already create a new table `ba775-team-4b.4b_dataset.different_categories_april_2020`-only use large categories-13 distinct categories\nCREATE MODEL\n model.users\nOPTIONS\n ( model_type = 'kmeans',\n num_clusters = 2,\n distance_type = 'euclidean') AS\nSELECT \nnum_of_accessories,num_of_apparel,num_of_appliances,num_of_auto,num_of_computers,num_of_construction,num_of_country_yard,num_of_electronics,num_of_furniture,num_of_kids,num_of_medicine,num_of_sport,num_of_stationery\nFROM `ba775-team-4b.4b_dataset.different_categories_april_2020`",
"_____no_output_____"
]
],
[
[
"# Tableau Dashboard \n\nhttps://prod-useast-a.online.tableau.com/t/soltaniehha/views/BA775Team4bProject_16040742108920/EcommerceDashboard?:origin=card_share_link&:embed=n",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image",
"_____no_output_____"
],
[
"# Top Purchase Brand of Feberary and April\nImage(filename='Top Brand.png')",
"_____no_output_____"
],
[
"# Abandon Rate of Feberary and April\nImage(filename='Abandon Rate.png')",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0b65b4d08fb3e7df8feb5f1518416f83ebaf6e1 | 10,225 | ipynb | Jupyter Notebook | stimuli/Play with Box2D.ipynb | jacobschenberg/block_construction | 079ee18de8023cddd0f4e361c6b891db63e9808e | [
"MIT"
] | null | null | null | stimuli/Play with Box2D.ipynb | jacobschenberg/block_construction | 079ee18de8023cddd0f4e361c6b891db63e9808e | [
"MIT"
] | null | null | null | stimuli/Play with Box2D.ipynb | jacobschenberg/block_construction | 079ee18de8023cddd0f4e361c6b891db63e9808e | [
"MIT"
] | null | null | null | 31.269113 | 112 | 0.579364 | [
[
[
"import sys\nimport importlib\nimport blockworld_helpers as utils\nfrom Box2D import *\nimport copy\nimport numpy as np",
"_____no_output_____"
],
[
"world = b2World(gravity=(0,-10), doSleep=True)\ngroundBody = world.CreateStaticBody(\n position=(0,-10),\n shapes=b2PolygonShape(box=(50,10)),\n )\n\nbody = world.CreateDynamicBody(position=(0,1))\nbox = body.CreatePolygonFixture(box=(1,1), density=1, friction=0.3)\nstart_positions = np.array([body.position for body in world.bodies])\n\ntimeStep = 1.0 / 60\n\nvel_iters, pos_iters = 6, 2\n\n# This is our little game loop.\nfor i in range(60):\n # Instruct the world to perform a single step of simulation. It is\n # generally best to keep the time step and iterations fixed.\n world.Step(timeStep, vel_iters, pos_iters)\n\n # Clear applied body forces. We didn't apply any forces, but you\n # should know about this function.\n world.ClearForces()\n \n # Now print the position and angle of the body.\n print(body.position, body.angle)\n\nend_positions = np.array([body.position for body in world.bodies])",
"b2Vec2(0,1.0045) 0.00011596875265240669\nb2Vec2(-1.51816e-08,1.01456) 0.00014852664025966078\nb2Vec2(-2.10134e-08,1.01463) 0.00011569393245736137\nb2Vec2(-1.64603e-08,1.01469) 9.021456935442984e-05\nb2Vec2(-1.25036e-08,1.01474) 7.045112579362467e-05\nb2Vec2(-9.7566e-09,1.01479) 5.5100925237638876e-05\nb2Vec2(-7.56404e-09,1.01482) 4.3170071876375005e-05\nb2Vec2(-5.88105e-09,1.01485) 3.387146716704592e-05\nb2Vec2(-4.65211e-09,1.01488) 2.6618676201906055e-05\nb2Vec2(-3.61407e-09,1.0149) 2.0953819330316037e-05\nb2Vec2(-2.75561e-09,1.01491) 1.6519050404895097e-05\nb2Vec2(-1.9739e-09,1.01493) 1.3042827958997805e-05\nb2Vec2(-1.49882e-09,1.01494) 1.0324913091608323e-05\nb2Vec2(-1.04999e-09,1.01495) 8.178866664820816e-06\nb2Vec2(-6.46138e-10,1.01496) 6.49100775262923e-06\nb2Vec2(-3.05554e-10,1.01496) 5.174590569367865e-06\nb2Vec2(-6.37775e-11,1.01497) 4.137596988584846e-06\nb2Vec2(4.73128e-11,1.01498) 3.3006342619046336e-06\nb2Vec2(6.02825e-11,1.01498) 2.6427705961395986e-06\nb2Vec2(1.32875e-10,1.01498) 2.1135940642125206e-06\nb2Vec2(2.12677e-10,1.01499) 1.698665755611728e-06\nb2Vec2(5.62766e-10,1.01499) 1.369299980069627e-06\nb2Vec2(7.63628e-10,1.01499) 1.1116081850559567e-06\nb2Vec2(8.19592e-10,1.01499) 8.969756208898616e-07\nb2Vec2(7.65913e-10,1.01499) 7.253673288687423e-07\nb2Vec2(7.91323e-10,1.01499) 5.965959530840337e-07\nb2Vec2(8.79194e-10,1.01499) 4.963729338669509e-07\nb2Vec2(1.09392e-09,1.015) 3.960239496336726e-07\nb2Vec2(1.2257e-09,1.015) 3.2436753372167004e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\nb2Vec2(1.20557e-09,1.015) 2.526756190945889e-07\n"
],
[
"epsilon = 0.1\n#start_positions = np.array([body.position for body in b2world_start.bodies])\n#end_positions = np.array([body.position for body in b2world_end.bodies])\nprint(start_positions)\nprint(end_positions)\nsum(sum(np.absolute(np.subtract(start_positions, end_positions))))\n)",
"[[ 0. -10.]\n [ 0. 1.]]\n[[ 0.00000000e+00 -1.00000000e+01]\n [ 1.20557464e-09 1.01499712e+00]]\n"
],
[
"print(len(world.bodies))",
"2\n"
],
[
"# Helper functions for interacting between stimulus generation and pybox2D\n\ndef b2_x(block):\n '''\n Takes a block from stimulus generation and returns the x value of the center of the block\n '''\n return ((block.x) + (block.width / 2))\n\ndef b2_y(block):\n '''\n Takes a block from stimulus generation and returns the y value of the center of the block\n '''\n return ((block.y) + (block.height / 2))\n \ndef add_block_to_world(block, b2world):\n '''\n Add block from stimulus generation to b2world\n '''\n body = b2world.CreateDynamicBody(position=(b2_x(block),b2_y(block)))\n world_block = body.CreatePolygonFixture(box=(block.width/2,block.height/2), density=1, friction=0.3)\n",
"_____no_output_____"
],
[
"importlib.reload(utils)\n\n# Create world for stability check\nb2world = b2World(gravity=(0,-10), doSleep=True)\ngroundBody = b2world.CreateStaticBody( #add ground\n position=(0,-10),\n shapes=b2PolygonShape(box=(50,10)),\n )\n\n# Add blocks\nworld = utils.World(world_width = 4,world_height = 4)\nworld.fill_world()\n\nfor block in world.blocks:\n b2block = add_block_to_world(block, b2world)\n ",
"_____no_output_____"
],
[
"b2world.bodies[1].fixtures[0].shape",
"_____no_output_____"
],
[
"# Run world\ntimeStep = 1.0 / 60\n\nvel_iters, pos_iters = 6, 2\n\n# This is our little game loop.\nfor i in range(60):\n # Instruct the world to perform a single step of simulation. It is\n # generally best to keep the time step and iterations fixed.\n b2world.Step(timeStep, vel_iters, pos_iters)\n\n # Clear applied body forces. We didn't apply any forces, but you\n # should know about this function.\n b2world.ClearForces()\n \n # Now print the position and angle of the body.\n print(world.blocks[1].x,world.blocks[1].y)\n print(b2world.bodies[2].position, body.angle)",
"_____no_output_____"
],
[
"b2world.bodies",
"_____no_output_____"
],
[
"importlib.reload(display_world)\ndisplay_world.display_world()",
"_____no_output_____"
],
[
"help(b2World)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b667bbc0faab33851de37119144c01f94bb7df | 355,943 | ipynb | Jupyter Notebook | Extra/Cusanovich_2018_subset/test_blacklist/run_umap_cusanovich2018subset_no_blacklist_filtering.ipynb | tAndreani/scATAC-benchmarking | 0b7167490aff986738985cdba688b48d4ed9988f | [
"MIT"
] | 2 | 2019-10-28T09:27:51.000Z | 2019-11-25T18:03:04.000Z | Extra/Cusanovich_2018_subset/test_blacklist/run_umap_cusanovich2018subset_no_blacklist_filtering.ipynb | josoga2/scATAC-benchmarking | 94118bbc0fb04c91f85c5bf4bc6a599ec706a218 | [
"MIT"
] | null | null | null | Extra/Cusanovich_2018_subset/test_blacklist/run_umap_cusanovich2018subset_no_blacklist_filtering.ipynb | josoga2/scATAC-benchmarking | 94118bbc0fb04c91f85c5bf4bc6a599ec706a218 | [
"MIT"
] | null | null | null | 465.284967 | 167,112 | 0.926879 | [
[
[
"library(data.table)\nlibrary(dplyr)\nlibrary(Matrix)\nlibrary(BuenColors)\nlibrary(stringr)\nlibrary(cowplot)\nlibrary(SummarizedExperiment)\nlibrary(chromVAR)\nlibrary(BSgenome.Hsapiens.UCSC.hg19)\nlibrary(JASPAR2016)\nlibrary(motifmatchr)\nlibrary(GenomicRanges)\nlibrary(irlba)\nlibrary(cicero)\nlibrary(umap)\nlibrary(cisTopic)\nlibrary(prabclus)\nlibrary(BrockmanR)\nlibrary(jackstraw)\nlibrary(RColorBrewer)",
"\nAttaching package: ‘dplyr’\n\nThe following objects are masked from ‘package:data.table’:\n\n between, first, last\n\nThe following objects are masked from ‘package:stats’:\n\n filter, lag\n\nThe following objects are masked from ‘package:base’:\n\n intersect, setdiff, setequal, union\n\nLoading required package: MASS\n\nAttaching package: ‘MASS’\n\nThe following object is masked from ‘package:dplyr’:\n\n select\n\nLoading required package: ggplot2\n\nAttaching package: ‘cowplot’\n\nThe following object is masked from ‘package:ggplot2’:\n\n ggsave\n\nLoading required package: GenomicRanges\nLoading required package: stats4\nLoading required package: BiocGenerics\nLoading required package: parallel\n\nAttaching package: ‘BiocGenerics’\n\nThe following objects are masked from ‘package:parallel’:\n\n clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,\n clusterExport, clusterMap, parApply, parCapply, parLapply,\n parLapplyLB, parRapply, parSapply, parSapplyLB\n\nThe following objects are masked from ‘package:Matrix’:\n\n colMeans, colSums, rowMeans, rowSums, which\n\nThe following objects are masked from ‘package:dplyr’:\n\n combine, intersect, setdiff, union\n\nThe following objects are masked from ‘package:stats’:\n\n IQR, mad, sd, var, xtabs\n\nThe following objects are masked from ‘package:base’:\n\n anyDuplicated, append, as.data.frame, basename, cbind, colMeans,\n colnames, colSums, dirname, do.call, duplicated, eval, evalq,\n Filter, Find, get, grep, grepl, intersect, is.unsorted, lapply,\n lengths, Map, mapply, match, mget, order, paste, pmax, pmax.int,\n pmin, pmin.int, Position, rank, rbind, Reduce, rowMeans, rownames,\n rowSums, sapply, setdiff, sort, table, tapply, union, unique,\n unsplit, which, which.max, which.min\n\nLoading required package: S4Vectors\n\nAttaching package: ‘S4Vectors’\n\nThe following object is masked from ‘package:Matrix’:\n\n expand\n\nThe following objects are masked from ‘package:dplyr’:\n\n first, rename\n\nThe following objects are masked from ‘package:data.table’:\n\n first, second\n\nThe following object is masked from ‘package:base’:\n\n expand.grid\n\nLoading required package: IRanges\n\nAttaching package: ‘IRanges’\n\nThe following objects are masked from ‘package:dplyr’:\n\n collapse, desc, slice\n\nThe following object is masked from ‘package:data.table’:\n\n shift\n\nLoading required package: GenomeInfoDb\nLoading required package: Biobase\nWelcome to Bioconductor\n\n Vignettes contain introductory material; view with\n 'browseVignettes()'. To cite Bioconductor, see\n 'citation(\"Biobase\")', and for packages 'citation(\"pkgname\")'.\n\nLoading required package: DelayedArray\nLoading required package: matrixStats\n\nAttaching package: ‘matrixStats’\n\nThe following objects are masked from ‘package:Biobase’:\n\n anyMissing, rowMedians\n\nThe following object is masked from ‘package:dplyr’:\n\n count\n\nLoading required package: BiocParallel\n\nAttaching package: ‘DelayedArray’\n\nThe following objects are masked from ‘package:matrixStats’:\n\n colMaxs, colMins, colRanges, rowMaxs, rowMins, rowRanges\n\nThe following objects are masked from ‘package:base’:\n\n aperm, apply\n\n\nLoading required package: BSgenome\nLoading required package: Biostrings\nLoading required package: XVector\n\nAttaching package: ‘Biostrings’\n\nThe following object is masked from ‘package:DelayedArray’:\n\n type\n\nThe following object is masked from ‘package:base’:\n\n strsplit\n\nLoading required package: rtracklayer\nLoading required package: monocle\nLoading required package: VGAM\nLoading required package: splines\nLoading required package: DDRTree\nLoading required package: Gviz\nLoading required package: grid\nWarning message:\n\"replacing previous import 'GenomicRanges::shift' by 'data.table::shift' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'data.table::last' by 'dplyr::last' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'GenomicRanges::union' by 'dplyr::union' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'GenomicRanges::intersect' by 'dplyr::intersect' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'GenomicRanges::setdiff' by 'dplyr::setdiff' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'data.table::first' by 'dplyr::first' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'data.table::between' by 'dplyr::between' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::failwith' by 'plyr::failwith' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::id' by 'plyr::id' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::summarize' by 'plyr::summarize' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::count' by 'plyr::count' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::desc' by 'plyr::desc' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::mutate' by 'plyr::mutate' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::arrange' by 'plyr::arrange' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::rename' by 'plyr::rename' when loading 'cisTopic'\"Warning message:\n\"replacing previous import 'dplyr::summarise' by 'plyr::summarise' when loading 'cisTopic'\"Loading required package: mclust\nPackage 'mclust' version 5.4.3\nType 'citation(\"mclust\")' for citing this R package in publications.\nLoading required package: tsne\n"
]
],
[
[
"#### define functions",
"_____no_output_____"
]
],
[
[
"read_FM <- function(filename){\n df_FM = data.frame(readRDS(filename),stringsAsFactors=FALSE,check.names=FALSE)\n rownames(df_FM) <- make.names(rownames(df_FM), unique=TRUE)\n df_FM[is.na(df_FM)] <- 0\n return(df_FM)\n}\n\nrun_pca <- function(mat,num_pcs=50,remove_first_PC=FALSE,scale=FALSE,center=FALSE){\n set.seed(2019) \n mat = as.matrix(mat)\n SVD = irlba(mat, num_pcs, num_pcs,scale=scale,center=center)\n sk_diag = matrix(0, nrow=num_pcs, ncol=num_pcs)\n diag(sk_diag) = SVD$d\n if(remove_first_PC){\n sk_diag[1,1] = 0\n SVD_vd = (sk_diag %*% t(SVD$v))[2:num_pcs,]\n }else{\n SVD_vd = sk_diag %*% t(SVD$v)\n }\n return(SVD_vd)\n}\n\nelbow_plot <- function(mat,num_pcs=50,scale=FALSE,center=FALSE,title='',width=3,height=3){\n set.seed(2019) \n mat = data.matrix(mat)\n SVD = irlba(mat, num_pcs, num_pcs,scale=scale,center=center)\n options(repr.plot.width=width, repr.plot.height=height)\n df_plot = data.frame(PC=1:num_pcs, SD=SVD$d);\n# print(SVD$d[1:num_pcs])\n p <- ggplot(df_plot, aes(x = PC, y = SD)) +\n geom_point(col=\"#cd5c5c\",size = 1) + \n ggtitle(title)\n return(p)\n}\n\nrun_umap <- function(fm_mat){\n umap_object = umap(t(fm_mat),random_state = 2019)\n df_umap = umap_object$layout\n return(df_umap)\n}\n\n\nplot_umap <- function(df_umap,labels,title='UMAP',colormap=colormap){\n set.seed(2019) \n df_umap = data.frame(cbind(df_umap,labels),stringsAsFactors = FALSE)\n colnames(df_umap) = c('umap1','umap2','celltype')\n df_umap$umap1 = as.numeric(df_umap$umap1)\n df_umap$umap2 = as.numeric(df_umap$umap2)\n options(repr.plot.width=4, repr.plot.height=4)\n p <- ggplot(shuf(df_umap), aes(x = umap1, y = umap2, color = celltype)) +\n geom_point(size = 1) + scale_color_manual(values = colormap) +\n ggtitle(title)\n return(p)\n}",
"_____no_output_____"
]
],
[
[
"### Input",
"_____no_output_____"
]
],
[
[
"workdir = '../output/'\n\npath_umap = paste0(workdir,'umap_rds/')\nsystem(paste0('mkdir -p ',path_umap))\n\npath_fm = paste0(workdir,'feature_matrices/')",
"_____no_output_____"
],
[
"metadata <- read.table('../input/metadata.tsv',\n header = TRUE,\n stringsAsFactors=FALSE,quote=\"\",row.names=1)",
"_____no_output_____"
],
[
"list.files(path_fm,pattern=\"^FM*\")",
"_____no_output_____"
],
[
"# read in feature matrices and double check if cell names of feature matrices are consistent with metadata\nflag_identical = c()\nfor (filename in list.files(path_fm,pattern=\"^FM*\")){\n filename_split = unlist(strsplit(sub('\\\\.rds$', '', filename),'_'))\n method_i = filename_split[2]\n if(method_i == 'chromVAR'){\n method_i = paste(filename_split[2],filename_split[4],sep='_')\n }\n print(paste0('Read in ','fm_',method_i))\n assign(paste0('fm_',method_i),read_FM(paste0(path_fm,filename)))\n #check if column names are the same\n flag_identical[[method_i]] = identical(colnames(eval(as.name(paste0('fm_',method_i)))),\n rownames(metadata))\n}",
"[1] \"Read in fm_SCRAT\"\n[1] \"Read in fm_SnapATAC\"\n"
],
[
"flag_identical",
"_____no_output_____"
],
[
"all(flag_identical)",
"_____no_output_____"
],
[
"labels = metadata$label\n\nnum_colors = length(unique(labels))\ncolormap = colorRampPalette(brewer.pal(8, \"Dark2\"))(num_colors)\nnames(colormap) = unique(metadata$label)",
"_____no_output_____"
],
[
"head(labels)",
"_____no_output_____"
]
],
[
[
"### SnapATAC",
"_____no_output_____"
]
],
[
[
"df_umap_SnapATAC <- run_umap(fm_SnapATAC)",
"_____no_output_____"
],
[
"head(df_umap_SnapATAC)",
"_____no_output_____"
],
[
"p_SnapATAC <- plot_umap(df_umap_SnapATAC,labels = labels,colormap = colormap,title='SnapATAC')\np_SnapATAC",
"_____no_output_____"
]
],
[
[
"### SCRAT",
"_____no_output_____"
]
],
[
[
"df_umap_SCRAT <- run_umap(fm_SCRAT)",
"_____no_output_____"
],
[
"p_SCRAT <- plot_umap(df_umap_SCRAT,labels = labels,colormap = colormap,title='SCRAT')\np_SCRAT",
"_____no_output_____"
]
],
[
[
"#### Save feature matrices and UMAP coordinates",
"_____no_output_____"
]
],
[
[
"dataset = 'cusanovich2018subset_no_blacklist_filtering'",
"_____no_output_____"
],
[
"saveRDS(df_umap_SnapATAC,paste0(path_umap,'df_umap_SnapATAC.rds'))\nsaveRDS(df_umap_SCRAT,paste0(path_umap,'df_umap_SCRAT.rds'))",
"_____no_output_____"
],
[
"save.image(file = 'run_umap_cusanovich2018subset_no_blacklist_filtering.RData')",
"_____no_output_____"
],
[
"fig_width = 8\nfig_height = 4\n\noptions(repr.plot.width=fig_width, repr.plot.height=fig_height)\ncombined_fig = cowplot::plot_grid(p_SnapATAC+theme(legend.position = \"none\"),\n p_SCRAT+theme(legend.position = \"none\"),\n labels = \"\",nrow = 1)",
"_____no_output_____"
],
[
"combined_fig",
"_____no_output_____"
],
[
"cowplot::ggsave(combined_fig,filename = \"Cusanovich_2018_ssubset_no_blacklist_filtering.pdf\", width = fig_width, height = fig_height)",
"_____no_output_____"
],
[
"cowplot::ggsave(p_SCRAT ,filename = \"cusanovich_legend.pdf\", width = fig_width, height = fig_height)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b66a7f601967c92c8d2dcb05a6a6957562ea81 | 227,133 | ipynb | Jupyter Notebook | nbs/052_learner.ipynb | clancy0614/tsai | 51dd8fbfc813e877536ab41ceb2da1b3ac84d9ca | [
"Apache-2.0"
] | 1 | 2021-11-15T07:44:29.000Z | 2021-11-15T07:44:29.000Z | nbs/052_learner.ipynb | nimingyonghuLiu/tsai | 577cb73373681a8dac46bcee0f23a24f0178b639 | [
"Apache-2.0"
] | null | null | null | nbs/052_learner.ipynb | nimingyonghuLiu/tsai | 577cb73373681a8dac46bcee0f23a24f0178b639 | [
"Apache-2.0"
] | 1 | 2021-11-14T02:58:25.000Z | 2021-11-14T02:58:25.000Z | 282.503731 | 159,060 | 0.914605 | [
[
[
"# default_exp learner",
"_____no_output_____"
]
],
[
[
"# Learner\n\n> This contains fastai Learner extensions.",
"_____no_output_____"
]
],
[
[
"#export\nfrom tsai.imports import *\nfrom tsai.data.core import *\nfrom tsai.data.validation import *\nfrom tsai.models.all import *\nfrom tsai.models.InceptionTimePlus import *\nfrom fastai.learner import * \nfrom fastai.vision.models.all import *\nfrom fastai.data.transforms import *",
"_____no_output_____"
],
[
"#export\n@patch\ndef show_batch(self:Learner, **kwargs):\n self.dls.show_batch(**kwargs)",
"_____no_output_____"
],
[
"# export\n \n@patch\ndef remove_all_cbs(self:Learner, max_iters=10):\n i = 0\n while len(self.cbs) > 0 and i < max_iters: \n self.remove_cbs(self.cbs)\n i += 1\n if len(self.cbs) > 0: print(f'Learner still has {len(self.cbs)} callbacks: {self.cbs}')",
"_____no_output_____"
],
[
"#export\n@patch\ndef one_batch(self:Learner, i, b): # this fixes a bug that will be managed in the next release of fastai\n self.iter = i\n# b_on_device = tuple( e.to(device=self.dls.device) for e in b if hasattr(e, \"to\")) if self.dls.device is not None else b\n b_on_device = to_device(b, device=self.dls.device) if self.dls.device is not None else b\n self._split(b_on_device)\n self._with_events(self._do_one_batch, 'batch', CancelBatchException)",
"_____no_output_____"
],
[
"#export\n@patch\ndef save_all(self:Learner, path='export', dls_fname='dls', model_fname='model', learner_fname='learner', verbose=False):\n path = Path(path)\n if not os.path.exists(path): os.makedirs(path)\n\n self.dls_type = self.dls.__class__.__name__\n if self.dls_type == \"MixedDataLoaders\":\n self.n_loaders = (len(self.dls.loaders), len(self.dls.loaders[0].loaders))\n dls_fnames = []\n for i,dl in enumerate(self.dls.loaders):\n for j,l in enumerate(dl.loaders):\n l = l.new(num_workers=1)\n torch.save(l, path/f'{dls_fname}_{i}_{j}.pth')\n dls_fnames.append(f'{dls_fname}_{i}_{j}.pth')\n else:\n dls_fnames = []\n self.n_loaders = len(self.dls.loaders)\n for i,dl in enumerate(self.dls):\n dl = dl.new(num_workers=1)\n torch.save(dl, path/f'{dls_fname}_{i}.pth')\n dls_fnames.append(f'{dls_fname}_{i}.pth')\n\n # Saves the model along with optimizer\n self.model_dir = path\n self.save(f'{model_fname}', with_opt=True)\n\n # Export learn without the items and the optimizer state for inference\n self.export(path/f'{learner_fname}.pkl')\n \n pv(f'Learner saved:', verbose)\n pv(f\"path = '{path}'\", verbose)\n pv(f\"dls_fname = '{dls_fnames}'\", verbose)\n pv(f\"model_fname = '{model_fname}.pth'\", verbose)\n pv(f\"learner_fname = '{learner_fname}.pkl'\", verbose)\n \n \ndef load_all(path='export', dls_fname='dls', model_fname='model', learner_fname='learner', device=None, pickle_module=pickle, verbose=False):\n\n if isinstance(device, int): device = torch.device('cuda', device)\n elif device is None: device = default_device()\n if device == 'cpu': cpu = True\n else: cpu = None\n\n path = Path(path)\n learn = load_learner(path/f'{learner_fname}.pkl', cpu=cpu, pickle_module=pickle_module)\n learn.load(f'{model_fname}', with_opt=True, device=device)\n\n \n if learn.dls_type == \"MixedDataLoaders\":\n dls_fnames = []\n _dls = []\n for i in range(learn.n_loaders[0]):\n _dl = []\n for j in range(learn.n_loaders[1]):\n l = torch.load(path/f'{dls_fname}_{i}_{j}.pth', map_location=device, pickle_module=pickle_module)\n l = l.new(num_workers=0)\n l.to(device)\n dls_fnames.append(f'{dls_fname}_{i}_{j}.pth')\n _dl.append(l)\n _dls.append(MixedDataLoader(*_dl, path=learn.dls.path, device=device, shuffle=l.shuffle))\n learn.dls = MixedDataLoaders(*_dls, path=learn.dls.path, device=device)\n\n else:\n loaders = []\n dls_fnames = []\n for i in range(learn.n_loaders):\n dl = torch.load(path/f'{dls_fname}_{i}.pth', map_location=device, pickle_module=pickle_module)\n dl = dl.new(num_workers=0)\n dl.to(device)\n first(dl)\n loaders.append(dl)\n dls_fnames.append(f'{dls_fname}_{i}.pth')\n learn.dls = type(learn.dls)(*loaders, path=learn.dls.path, device=device)\n\n\n pv(f'Learner loaded:', verbose)\n pv(f\"path = '{path}'\", verbose)\n pv(f\"dls_fname = '{dls_fnames}'\", verbose)\n pv(f\"model_fname = '{model_fname}.pth'\", verbose)\n pv(f\"learner_fname = '{learner_fname}.pkl'\", verbose)\n return learn\n\nload_learner_all = load_all",
"_____no_output_____"
],
[
"#export\n@patch\n@delegates(subplots)\ndef plot_metrics(self: Recorder, nrows=None, ncols=None, figsize=None, final_losses=True, perc=.5, **kwargs):\n n_values = len(self.recorder.values)\n if n_values < 2: \n print('not enough values to plot a chart')\n return\n metrics = np.stack(self.values)\n n_metrics = metrics.shape[1]\n names = self.metric_names[1:n_metrics+1]\n if final_losses: \n sel_idxs = int(round(n_values * perc))\n if sel_idxs >= 2:\n metrics = np.concatenate((metrics[:,:2], metrics), -1)\n names = names[:2] + names\n else: \n final_losses = False\n n = len(names) - 1 - final_losses\n if nrows is None and ncols is None:\n nrows = int(math.sqrt(n))\n ncols = int(np.ceil(n / nrows))\n elif nrows is None: nrows = int(np.ceil(n / ncols))\n elif ncols is None: ncols = int(np.ceil(n / nrows))\n figsize = figsize or (ncols * 6, nrows * 4)\n fig, axs = subplots(nrows, ncols, figsize=figsize, **kwargs)\n axs = [ax if i < n else ax.set_axis_off() for i, ax in enumerate(axs.flatten())][:n]\n axs = ([axs[0]]*2 + [axs[1]]*2 + axs[2:]) if final_losses else ([axs[0]]*2 + axs[1:])\n for i, (name, ax) in enumerate(zip(names, axs)):\n if i in [0, 1]:\n ax.plot(metrics[:, i], color='#1f77b4' if i == 0 else '#ff7f0e', label='valid' if i == 1 else 'train')\n ax.set_title('losses')\n ax.set_xlim(0, len(metrics)-1)\n elif i in [2, 3] and final_losses:\n ax.plot(np.arange(len(metrics) - sel_idxs, len(metrics)), metrics[-sel_idxs:, i], \n color='#1f77b4' if i == 2 else '#ff7f0e', label='valid' if i == 3 else 'train')\n ax.set_title('final losses')\n ax.set_xlim(len(metrics) - sel_idxs, len(metrics)-1)\n # ax.set_xticks(np.arange(len(metrics) - sel_idxs, len(metrics)))\n else:\n ax.plot(metrics[:, i], color='#1f77b4' if i == 0 else '#ff7f0e', label='valid' if i > 0 else 'train')\n ax.set_title(name if i >= 2 * (1 + final_losses) else 'losses')\n ax.set_xlim(0, len(metrics)-1)\n ax.legend(loc='best')\n ax.grid(color='gainsboro', linewidth=.5)\n plt.show()\n \n \n@patch\n@delegates(subplots)\ndef plot_metrics(self: Learner, **kwargs):\n self.recorder.plot_metrics(**kwargs)",
"_____no_output_____"
],
[
"#export\n@patch\n@delegates(subplots)\ndef show_probas(self:Learner, figsize=(6,6), ds_idx=1, dl=None, one_batch=False, max_n=None, **kwargs):\n recorder = copy(self.recorder) # This is to avoid loss of recorded values while generating preds\n if one_batch: dl = self.dls.one_batch()\n probas, targets = self.get_preds(ds_idx=ds_idx, dl=[dl] if dl is not None else None)\n if probas.ndim == 2 and probas.min() < 0 or probas.max() > 1: probas = nn.Softmax(-1)(probas)\n if not isinstance(targets[0].item(), Integral): return\n targets = targets.flatten()\n if max_n is not None:\n idxs = np.random.choice(len(probas), max_n, False)\n probas, targets = probas[idxs], targets[idxs]\n fig = plt.figure(figsize=figsize, **kwargs)\n classes = np.unique(targets)\n nclasses = len(classes)\n vals = np.linspace(.5, .5 + nclasses - 1, nclasses)[::-1]\n plt.vlines(.5, min(vals) - 1, max(vals), color='black', linewidth=.5)\n cm = plt.get_cmap('gist_rainbow')\n color = [cm(1.* c/nclasses) for c in range(1, nclasses + 1)][::-1]\n class_probas = np.array([probas[i,t] for i,t in enumerate(targets)])\n for i, c in enumerate(classes):\n plt.scatter(class_probas[targets == c] if nclasses > 2 or i > 0 else 1 - class_probas[targets == c],\n targets[targets == c] + .5 * (np.random.rand((targets == c).sum()) - .5), color=color[i], edgecolor='black', alpha=.2, s=100)\n if nclasses > 2: plt.vlines((targets == c).float().mean(), i - .5, i + .5, color='r', linewidth=.5)\n plt.hlines(vals, 0, 1)\n plt.ylim(min(vals) - 1, max(vals))\n plt.xlim(0,1)\n plt.xticks(np.linspace(0,1,11), fontsize=12)\n plt.yticks(classes, [self.dls.vocab[x] for x in classes], fontsize=12)\n plt.title('Predicted proba per true class' if nclasses > 2 else 'Predicted class 1 proba per true class', fontsize=14)\n plt.xlabel('Probability', fontsize=12)\n plt.ylabel('True class', fontsize=12)\n plt.grid(axis='x', color='gainsboro', linewidth=.2)\n plt.show()\n self.recorder = recorder",
"_____no_output_____"
],
[
"#export \nall_archs = [FCN, FCNPlus, InceptionTime, InceptionTimePlus, InCoordTime, XCoordTime,\n InceptionTimePlus17x17, InceptionTimePlus32x32, InceptionTimePlus47x47, InceptionTimePlus62x62,\n InceptionTimeXLPlus, MultiInceptionTimePlus, MiniRocketClassifier, MiniRocketRegressor, \n MiniRocketVotingClassifier, MiniRocketVotingRegressor, MiniRocketFeaturesPlus, MiniRocketPlus, MiniRocketHead,\n InceptionRocketFeaturesPlus, InceptionRocketPlus, MLP, MultiInputNet, OmniScaleCNN, RNN, LSTM, GRU, RNNPlus, LSTMPlus, GRUPlus, \n RNN_FCN, LSTM_FCN, GRU_FCN, MRNN_FCN, MLSTM_FCN, MGRU_FCN, ROCKET, RocketClassifier, RocketRegressor, ResCNNBlock, ResCNN, \n ResNet, ResNetPlus, TCN, TSPerceiver, TST, TSTPlus, MultiTSTPlus, TSiTPlus, TSiT, InceptionTSiTPlus, InceptionTSiT, \n TabFusionTransformer, TSTabFusionTransformer, TabModel, TabTransformer, TransformerModel, XCM, XCMPlus, \n xresnet1d18, xresnet1d34, xresnet1d50, xresnet1d101, xresnet1d152, xresnet1d18_deep,\n xresnet1d34_deep, xresnet1d50_deep, xresnet1d18_deeper, xresnet1d34_deeper, xresnet1d50_deeper,\n XResNet1dPlus, xresnet1d18plus, xresnet1d34plus, xresnet1d50plus, xresnet1d101plus,\n xresnet1d152plus, xresnet1d18_deepplus, xresnet1d34_deepplus, xresnet1d50_deepplus,\n xresnet1d18_deeperplus, xresnet1d34_deeperplus, xresnet1d50_deeperplus, XceptionTime, XceptionTimePlus\n ]\n\nall_archs_names = [arch.__name__ for arch in all_archs]\ndef get_arch(arch_name):\n assert arch_name in all_archs_names, \"confirm the name of the architecture\"\n idx = all_archs_names.index(arch_name)\n return all_archs[idx]",
"_____no_output_____"
],
[
"arch_name = 'InceptionTimePlus'\ntest_eq(get_arch('InceptionTimePlus').__name__, arch_name)",
"_____no_output_____"
],
[
"#export\n@delegates(build_ts_model)\ndef ts_learner(dls, arch=None, c_in=None, c_out=None, seq_len=None, d=None, splitter=trainable_params,\n # learner args\n loss_func=None, opt_func=Adam, lr=defaults.lr, cbs=None, metrics=None, path=None,\n model_dir='models', wd=None, wd_bn_bias=False, train_bn=True, moms=(0.95,0.85,0.95),\n # other model args\n **kwargs):\n\n if arch is None: arch = InceptionTimePlus\n elif isinstance(arch, str): arch = get_arch(arch)\n model = build_ts_model(arch, dls=dls, c_in=c_in, c_out=c_out, seq_len=seq_len, d=d, **kwargs)\n try:\n model[0], model[1]\n subscriptable = True\n except:\n subscriptable = False\n if subscriptable: splitter = ts_splitter\n if loss_func is None:\n if hasattr(dls, 'loss_func'): loss_func = dls.loss_func\n elif hasattr(dls, 'train_ds') and hasattr(dls.train_ds, 'loss_func'): loss_func = dls.train_ds.loss_func\n elif hasattr(dls, 'cat') and not dls.cat: loss_func = MSELossFlat()\n \n learn = Learner(dls=dls, model=model,\n loss_func=loss_func, opt_func=opt_func, lr=lr, cbs=cbs, metrics=metrics, path=path, splitter=splitter,\n model_dir=model_dir, wd=wd, wd_bn_bias=wd_bn_bias, train_bn=train_bn, moms=moms, )\n\n # keep track of args for loggers\n store_attr('arch', self=learn)\n\n return learn",
"_____no_output_____"
],
[
"#export\n@delegates(build_tsimage_model)\ndef tsimage_learner(dls, arch=None, pretrained=False,\n # learner args\n loss_func=None, opt_func=Adam, lr=defaults.lr, cbs=None, metrics=None, path=None,\n model_dir='models', wd=None, wd_bn_bias=False, train_bn=True, moms=(0.95,0.85,0.95),\n # other model args\n **kwargs):\n\n if arch is None: arch = xresnet34\n elif isinstance(arch, str): arch = get_arch(arch)\n model = build_tsimage_model(arch, dls=dls, pretrained=pretrained, **kwargs)\n learn = Learner(dls=dls, model=model,\n loss_func=loss_func, opt_func=opt_func, lr=lr, cbs=cbs, metrics=metrics, path=path,\n model_dir=model_dir, wd=wd, wd_bn_bias=wd_bn_bias, train_bn=train_bn, moms=moms)\n\n # keep track of args for loggers\n store_attr('arch', self=learn)\n\n return learn",
"_____no_output_____"
],
[
"#export\n@patch\ndef decoder(self:Learner, o): return L([self.dls.decodes(oi) for oi in o])",
"_____no_output_____"
],
[
"# export\n\n@patch\n@delegates(GatherPredsCallback.__init__)\ndef get_X_preds(self: Learner, X, y=None, bs=64, with_input=False, with_decoded=True, with_loss=False, **kwargs):\n if with_loss and y is None:\n print('cannot find loss as y=None')\n with_loss = False\n dl = self.dls.valid.new_dl(X, y=y)\n dl.bs = ifnone(bs, self.dls.bs)\n output = list(self.get_preds(dl=dl, with_input=with_input, with_decoded=with_decoded, with_loss=with_loss, reorder=False))\n if with_decoded and hasattr(self.dls, 'vocab'):\n output[2 + with_input] = L([self.dls.vocab[p] for p in output[2 + with_input]])\n return tuple(output)",
"_____no_output_____"
],
[
"from tsai.data.all import *\nfrom tsai.data.core import *\nfrom tsai.models.FCNPlus import *\ndsid = 'OliveOil'\nX, y, splits = get_UCR_data(dsid, verbose=True, split_data=False)\ntfms = [None, [Categorize()]]\ndls = get_ts_dls(X, y, splits=splits, tfms=tfms)\nlearn = ts_learner(dls, FCNPlus)\nfor p in learn.model.parameters():\n p.requires_grad=False\ntest_eq(count_parameters(learn.model), 0)\nlearn.freeze()\ntest_eq(count_parameters(learn.model), 1540)\nlearn.unfreeze()\ntest_eq(count_parameters(learn.model), 264580)\n\nlearn = ts_learner(dls, 'FCNPlus')\nfor p in learn.model.parameters():\n p.requires_grad=False\ntest_eq(count_parameters(learn.model), 0)\nlearn.freeze()\ntest_eq(count_parameters(learn.model), 1540)\nlearn.unfreeze()\ntest_eq(count_parameters(learn.model), 264580)",
"Dataset: OliveOil\nX : (60, 1, 570)\ny : (60,)\nsplits : (#30) [0,1,2,3,4,5,6,7,8,9...] (#30) [30,31,32,33,34,35,36,37,38,39...] \n\n"
],
[
"learn.show_batch();",
"_____no_output_____"
],
[
"learn.fit_one_cycle(2, lr_max=1e-3)",
"_____no_output_____"
],
[
"dsid = 'OliveOil'\nX, y, splits = get_UCR_data(dsid, split_data=False)\ntfms = [None, [Categorize()]]\ndls = get_ts_dls(X, y, tfms=tfms, splits=splits)\nlearn = ts_learner(dls, FCNPlus, metrics=accuracy)\nlearn.fit_one_cycle(2)\nlearn.plot_metrics()\nlearn.show_probas()",
"_____no_output_____"
],
[
"learn.save_all()\ndel learn\nlearn = load_all()",
"_____no_output_____"
],
[
"test_probas, test_targets, test_preds = learn.get_X_preds(X[0:10], with_decoded=True)\ntest_probas, test_targets, test_preds",
"_____no_output_____"
],
[
"test_probas2, test_targets2, test_preds2 = learn.get_X_preds(X[0:10], y[0:10], with_decoded=True)\ntest_probas2, test_targets2, test_preds2",
"_____no_output_____"
],
[
"test_eq(test_probas, test_probas2)\ntest_eq(test_preds, test_preds2)",
"_____no_output_____"
],
[
"learn.fit_one_cycle(1, lr_max=1e-3)",
"_____no_output_____"
],
[
"#hide\nfrom tsai.imports import create_scripts\nfrom tsai.export import get_nb_name\nnb_name = get_nb_name()\ncreate_scripts(nb_name);",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b66b0fdbdcdc67616ec48ac2b933eb0690c41f | 196,224 | ipynb | Jupyter Notebook | _notebooks/2020-10-23-Stock_Price_Indicators_using_the_python_FINTA_library.ipynb | IjeomaOdoko/my-blog | 6d0dea6c4aeecc079c02bd407e3fc24f677ee396 | [
"Apache-2.0"
] | null | null | null | _notebooks/2020-10-23-Stock_Price_Indicators_using_the_python_FINTA_library.ipynb | IjeomaOdoko/my-blog | 6d0dea6c4aeecc079c02bd407e3fc24f677ee396 | [
"Apache-2.0"
] | 2 | 2021-09-28T05:34:06.000Z | 2022-02-26T10:01:11.000Z | _notebooks/2020-10-23-Stock_Price_Indicators_using_the_python_FINTA_library.ipynb | IjeomaOdoko/my-blog | 6d0dea6c4aeecc079c02bd407e3fc24f677ee396 | [
"Apache-2.0"
] | null | null | null | 92.689655 | 86,907 | 0.680977 | [
[
[
"# \"Price Charts with Technical Indicators\"\n\n> \"Calculating Stock Price Indicators using FINTA python library, and visualizing using plotly python library.\"\n\n- toc: false\n- branch: master\n- badges: true\n- comments: true\n- author: Ijeoma Odoko\n- categories: [stocks, python, finta, pandas, plotly, ipywidgets]\n\n",
"_____no_output_____"
],
[
"\n\nImage credits to Markus Spiske - Unsplash photos\n\n\n",
"_____no_output_____"
],
[
"## About ",
"_____no_output_____"
],
[
"\nThere are a couple of libraries to use to calculate technical indicators for stocks. In previous posts, we had tried out the following python libraries: \n\n> mplfinance\n\n> TA-lib\n\nIn this post we will be looking at the [finta library](https://https://pypi.org/project/finta/). ",
"_____no_output_____"
],
[
"## Install required dependencies on Google Colab \n\n",
"_____no_output_____"
]
],
[
[
"!pip install finta",
"Requirement already satisfied: finta in /usr/local/lib/python3.6/dist-packages (1.2)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from finta) (1.18.5)\nRequirement already satisfied: pandas in /usr/local/lib/python3.6/dist-packages (from finta) (1.1.2)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.6/dist-packages (from pandas->finta) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas->finta) (2018.9)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.7.3->pandas->finta) (1.15.0)\n"
]
],
[
[
"## Load required libraries\n\n",
"_____no_output_____"
]
],
[
[
"from pandas_datareader import data\nimport numpy as np\nfrom datetime import datetime \nfrom datetime import date, timedelta\nimport pandas as pd\n\n# import ipwidgets library and functions\nfrom __future__ import print_function\nfrom ipywidgets import interact, interactive, fixed\nimport ipywidgets as widgets\nfrom IPython.display import display\n\nfrom finta import TA",
"_____no_output_____"
]
],
[
[
"## Create widgets and dataframe for the stock data\n\n\n*Instructions for use. *\n1. Insert tuple of stock list. \n2. Select stock from dropdown. \n3. Select number of calendar days for dates from the last trading day. \n4. Rerun all code after.",
"_____no_output_____"
]
],
[
[
"# Insert a tuple of unique tickers into the options variables.\n#tickers = ('MMM', 'AOS', 'AAN', 'ABB', 'ABT', 'ABBV', 'ABM', 'ACN', 'AYI', 'GOLF', 'ADCT', 'ADT', 'AAP', 'ADSW', 'WMS', 'ACM', 'AEG', 'AER', 'AJRD', 'AMG', 'AFL', 'AGCO', 'A', 'AEM', 'ADC', 'AL', 'APD', 'AGI', 'ALK', 'ALB', 'ACI', 'AA', 'ALC', 'ARE', 'AQN', 'BABA', 'Y')\n#tickers = ('ARKF', 'ARKG', 'ARKK', 'ARKW', 'QQQ','TQQQ', 'VCR', \"KARS\", 'ZNGA')\ntickers = ('SOXX', 'SOXL', 'TQQQ', 'QQQ', 'ARKK', 'ARKW', 'FDN', 'XLY', 'VCR', 'FPX', 'SMH')",
"_____no_output_____"
],
[
"# create dropdown for selected stocks\nstock_ticker = widgets.Dropdown(\n options= tickers,\n description='Select Stock Ticker',\n disabled=False,\n style = {'description_width': 'initial'}, \n layout = {'width': '200px'}\n)\n\n# create selection slider for days\nw = widgets.IntSlider(\n value=90,\n min=5,\n max=365,\n step=1,\n description = 'Calendar days',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d',\n style = {'description_width': 'initial','handle_color' : 'blue'}, \n layout = {'width': '400px'}\n)\n\n# create function for time frame of selected calendar days from today\ndef timeframe(w):\n days = timedelta(w)\n start = date.today() - days\n today = date.today()\n print('Start Date: ',start, ' ' ,'Last Date: ',today)\n \ndates = widgets.interactive_output(timeframe, {'w': w} )\n\ndisplay(stock_ticker, w, dates)",
"_____no_output_____"
]
],
[
[
"## Download data for the stock",
"_____no_output_____"
]
],
[
[
"# create text to show stock ticker \n\nv = widgets.Text(\n value=stock_ticker.value,\n description='Stockticker:',\n disabled=True\n)\n\n# create function to load stock data from yahoo \ndef load_stock_data(stock_ticker, w):\n start = date.today() - timedelta(w)\n today = date.today()\n stock_data = data.DataReader(stock_ticker, start=start, end=today, data_source='yahoo')\n return stock_data\n\n# create dataframe for selected stock\nstock = load_stock_data(stock_ticker.value, w.value)\n\n# display ticker and dataframe\n\ndisplay(v, stock)",
"_____no_output_____"
],
[
"# format dataframe in the format required by finta \nohlcv = stock[['Open', 'High', 'Low', 'Close', 'Volume']] # select the columns in the order required\n\nohlcv.columns = ['open', 'high', 'low', 'close', 'volume'] # rename the columns\n\nohlcv",
"_____no_output_____"
]
],
[
[
"## Calculate some Stock Price Indicators",
"_____no_output_____"
]
],
[
[
"# create example dataframe to try out the functions\n\nex_df = ohlcv.copy()\n\nex_df['RSI'] = TA.RSI(ex_df)\nex_df['Simple_Moving_Average_50'] = TA.SMA(ex_df, 50)\nex_df[['macd', 'macd_s']] = TA.MACD(ex_df)\n\nex_df",
"_____no_output_____"
]
],
[
[
"## Create a function to create a dataframe that captures some stock technical indicators",
"_____no_output_____"
]
],
[
[
"# create function to create the Stock Indicator dataframe \n\ndef create_dataframe(df):\n\n \"\"\"\n This function creates a Dataframe for key indicators\n \"\"\"\n \n df['Daily_Returns'] = df['close'].pct_change() # create column for daily returns\n df['Price_Up_or_Down'] = np.where(df['Daily_Returns'] < 0, -1, 1) # create column for price up or down \n \n # add columns for the volatility and volume indicators\n \n df['Average_True_Range'] = TA.ATR(df)\n df['On_Balance_Volume'] = TA.OBV(df)\n df['Volume_Flow_Indicator'] = TA.VFI(df)\n\n ## add column for moving averages\n \n df['Simple_Moving_Average_50'] = TA.SMA(df, 50)\n #df['Simple_Moving_Average_200'] = TA.SMA(df, 200)\n df['Volume Weighted Average Price'] = TA.VWAP(df)\n df['Exponential_Moving_Average_50'] = TA.EMA(df, 50)\n\n # add columns for momentum indicators\n \n df['ADX'] = TA.ADX(df) #create column for ADX assume timeperiod of 14 days\n df['RSI'] = TA.RSI(df) #create column for RSI assume timeperiod of 14 days \n df['William %R'] = TA.WILLIAMS(df) #create column for William %R use high, low and close, and assume timeperiod of 14 days\n df['MFI'] = TA.MFI(df) #create column for MFI use high, low and close, and assume timeperiod of 14 days\n df['MOM'] = TA.MOM(df)\n df[['macd', 'macd_signal']] = TA.MACD(df)\n\n return df # return the dataframe",
"_____no_output_____"
],
[
"# Create a dataframe with all the stock indicators you indicated in the create dataframe function\n\nstocks_df = create_dataframe(df = ohlcv)\n\nstocks_df",
"_____no_output_____"
]
],
[
[
"## VISUALIZATIONS USING PLOTLY",
"_____no_output_____"
],
[
"## Price Action Chart",
"_____no_output_____"
]
],
[
[
"# create OHLC charts with Plotly \n\nimport plotly.graph_objects as go\n\n\nfig_ohlc = go.Figure(data=[go.Ohlc(x=stocks_df.index,\n open=stocks_df['open'],\n high=stocks_df['high'],\n low=stocks_df['low'],\n close=stocks_df['close'], showlegend=False)])\n \nfig_ohlc.update_layout(title = 'Price Action Chart', yaxis_title = 'Stock Price', template = 'presentation')\n\nfig_ohlc.update(layout_xaxis_rangeslider_visible=False)\n\ndisplay(v)\n\nfig_ohlc.show()",
"_____no_output_____"
],
[
"# create Candlestick charts with Plotly \n\nimport plotly.graph_objects as go\n\n\nfig_candle = go.Figure(data=[go.Candlestick(x=stocks_df.index,\n open=stocks_df['open'],\n high=stocks_df['high'],\n low=stocks_df['low'],\n close=stocks_df['close'], showlegend=False)])\n \nfig_candle.update_layout(title = 'Price Action Chart', yaxis_title = 'Stock Price', template = 'presentation')\n\nfig_candle.update(layout_xaxis_rangeslider_visible=False)\n\ndisplay(v)\n\nfig_candle.show()",
"_____no_output_____"
]
],
[
[
"## Momentum Indicators",
"_____no_output_____"
]
],
[
[
"#hide\nimport plotly.graph_objects as go\nimport plotly.offline as pyo\n\ntrace1 = go.Scatter(x=stocks_df.index, y=stocks_df['macd'], mode='lines', marker=dict(color=\"green\"), showlegend=True, name='macd')\n\ntrace2 = go.Scatter(x=stocks_df.index, y=stocks_df['macd_signal'], mode='lines', marker=dict(color=\"blue\"), showlegend=True, name='macd_signal')\n\ndata= [trace1, trace2]\n\nlayout = go.Layout(title = 'MACD indicator')\n\nfig = go.Figure(data=data, layout=layout)\npyo.plot(fig, filename='MACD_indicator.html')",
"_____no_output_____"
],
[
"import plotly.graph_objects as go\n\ntrace1 = go.Scatter(x=stocks_df.index, y=stocks_df['macd'], mode='lines', marker=dict(color=\"green\"), showlegend=True, name='macd')\n\ntrace2 = go.Scatter(x=stocks_df.index, y=stocks_df['macd_signal'], mode='lines', marker=dict(color=\"blue\"), showlegend=True, name='macd_signal')\n\ndata= [trace1, trace2]\n\nlayout = go.Layout(title = 'MACD indicator')\n\nfig = go.Figure(data=data, layout=layout)\nfig.show()",
"_____no_output_____"
]
],
[
[
"# References \n\n[Plotly Figure Reference](https://plotly.com/python/reference/index/) accessed October 22, 2020.\n\n[FinTA (Financial Technical Analysis) python library](https://github.com/peerchemist/finta) accessed October 22, 2020.\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d0b66d0dfa3f990b0fd96fa6289a3ccf59fda428 | 11,435 | ipynb | Jupyter Notebook | manuscript_results/motivating_example_high_corr.ipynb | lwa19/susie-paper | 9eb53b2bb05ad65714b059e5af94802975f66f58 | [
"MIT"
] | 3 | 2021-02-12T20:10:46.000Z | 2021-11-10T19:30:37.000Z | manuscript_results/motivating_example_high_corr.ipynb | lwa19/susie-paper | 9eb53b2bb05ad65714b059e5af94802975f66f58 | [
"MIT"
] | 9 | 2019-12-04T03:14:11.000Z | 2022-02-09T22:40:21.000Z | manuscript_results/motivating_example_high_corr.ipynb | lwa19/susie-paper | 9eb53b2bb05ad65714b059e5af94802975f66f58 | [
"MIT"
] | 4 | 2018-12-29T05:45:15.000Z | 2021-11-10T21:46:31.000Z | 44.150579 | 399 | 0.606034 | [
[
[
"# Experiment with variables of given high correlation structure\n\nThis notebook is meant to address to a shared concern from two referees. The [motivating example](motivating_example.html) in the manuscript was designed to be a simple toy for illustrating the novel type of inference SuSiE offers. Here are some slightly more complicated examples, based on the motivating example, but with variables in high (rather than perfect) correlations with each other.",
"_____no_output_____"
],
[
"## $x_1$ and $x_2$ are highly correlated",
"_____no_output_____"
],
[
"Following a reviewer's\nsuggestion, we simulated two variables, $x_1$ and $x_2$, with high but\nnot perfect correlation ($0.9$). Specifically, we simulated $n = 600$\nsamples stored as an $X_{600 \\times 2}$ matrix, in which each row was\ndrawn *i.i.d.* from a normal distribution with mean zero and\n$\\mathrm{cor}(x_1, x_2) = 0.9$. \n\nWe then simulated $y_i = x_{i1} \\beta_1 + x_{i2} \\beta_2 + \\varepsilon_i$, \nwith $\\beta_1 = 1, \\beta_2 = 0$,\nand $\\varepsilon_i$ *i.i.d.* normal with zero mean and standard\ndeviation of 3. We performed 1,000 replicates of this simulation\n(generated with different random number seeds).",
"_____no_output_____"
],
[
"In this simulation, the correlation between $x_1$ and $x_2$ is still\nsufficiently high (0.9) to make distinguishing between the two\nvariables somewhat possible, but not non entirely straightforward. For\nexample, when we run lasso (using `cv.glmnet` from the `glmnet`\nR package) on these data it wrongly selected $x_2$ as having\nnon-zero coefficient in about 10% of the simulations (95 out of\n1,000), and correctly selected $x_1$ in about 96% of simulations (956\nout of 1,000). Note that the lasso does not assess uncertainty in\nvariable selection, so these results are not directly comparable\nwith SuSiE CSs below; however, the lasso results demonstrate that\ndistinguishing the correct variable here is possible, but not so easy\nthat the example is uninteresting.",
"_____no_output_____"
],
[
"Ideally, then, SuSiE should identify variable $x_1$ as an effect\nvariable and drop $x_2$ as often as possible. However, due to the high\ncorrelation between the variables, it is inevitable that some\n95% SuSiE credible sets (CS) will also contain $x_2$. Most\nimportant is that we should avoid, as much as possible, reporting a CS\nthat contains *only* $x_2$, since the goal is that 95% of CSs\nshould contain at least one effect variable. The SuSiE results (SuSiE version 0.9.1 on R 3.5.2) \nare summarized below. The code used for the simulation [can be found here](https://github.com/stephenslab/susie-paper/blob/master/src/ref_3_question.R).",
"_____no_output_____"
],
[
"| CSs | count |\n| :---- | ----: |\n| (1) | 829 |\n| (1,2) | 169 |\n| **(2)** | 2 |",
"_____no_output_____"
],
[
"Highlighted in **bold** are CSs that do *not* contain\nthe true effect variable --- there are 2 of them out of 1,000 CSs\ndetected. In summary, SuSiE precisely identifies the effect\nvariable in a single CS in the majority (83%) of the simulations, and\nprovides a \"valid\" CS (*i.e.*, one containing an effect\nvariable) in almost all simulations (998 out of 1,000). Further, even\nwhen SuSiE reports a CS including both variables, it consistently\nassigns higher posterior inclusion probability (PIP) to the correct\nvariable, $x_1$: among the 169 CSs that contain both variables, the\nmedian PIPs for $x_1$ and $x_2$ were 0.86 and 0.14, respectively.",
"_____no_output_____"
],
[
"## When an additional non-effect variable highly correlated with both variable groups",
"_____no_output_____"
],
[
"Another referee suggested the following:\n\n> Suppose\nwe have another predictor $x_5$, which is both correlated with $(x_1,\nx_2)$ and $(x_3, x_4)$. Say $\\mathrm{cor}(x_1, x_5) = 0.9$,\n$\\mathrm{cor}(x_2, x_5) = 0.7$, and $\\mathrm{cor}(x_5, x_3)\n= \\mathrm{cor}(x_5, x_4) = 0.8$. Does the current method assign $x_5$\nto the $(x_1, x_2)$ group or the $(x_3, x_4)$ group?",
"_____no_output_____"
],
[
"Following the suggestion, we simulated $x_1, \\ldots, x_5$ from a\nmultivariate normal with zero mean and the covariance matrix\napproximately as given in Table below. (Since this matrix is\nnot quite positive definite, in our R code we used `nearPD` from\nthe `Matrix` package to generate the nearest positive definite\nmatrix --- the entries of the resulting covariance matrix differ only\nvery slightly from those in Table below, with a maximum\nabsolute difference of 0.0025 between corresponding elements in the\ntwo matrices).\n\n| | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ |\n| ------: | ------: | ------: | ------: | ------: | ------: |\n| $x_1$ | 1.00 | 0.92 | 0.70 | 0.70 | 0.90 |\n| $x_2$ | 0.92 | 1.00 | 0.70 | 0.70 | 0.70 |\n| $x_3$ | 0.70 | 0.70 | 1.00 | 0.92 | 0.80 |\n| $x_4$ | 0.70 | 0.70 | 0.92 | 1.00 | 0.80 |\n| $x_5$ | 0.90 | 0.70 | 0.80 | 0.80 | 1.00 |\n",
"_____no_output_____"
],
[
"We simulated $n = 600$ samples from this\nmultivariate normal distribution, then we simulated $n = 600$\nresponses $y_i$ from the regression model $y_i = x_{i1} \\beta_1 + \\cdots x_{i5} \\beta_5 + \\varepsilon_i$, \nwith $\\beta = (0, 1, 1, 0, 0)^T$, and $\\varepsilon_i$ *i.i.d.* normal with zero mean and\nstandard deviation of 3. We repeated this simulation procedure 1,000\ntimes with different random seeds, and each time we fit a SuSiE\nmodel to the simulated data by running the IBSS algorithm. To\nsimplify the example, we ran the IBSS algorithm with $L = 2$, and\nfixed the $\\sigma_0^2 = 1$. Similar results were obtained when we used\nlarger values of $L$, and when $\\sigma_0^2$ was estimated. For more\ndetails on how the data were simulated and how the SuSiE models\nwere fitted to the data sets, [see this script](https://github.com/stephenslab/susie-paper/blob/master/src/ref_4_question.R).",
"_____no_output_____"
],
[
"Like the toy motivating example given in the paper, in this simulation\nthe first two predictors are strongly correlated with each other, so\nit may be difficult to distinguish among them, and likewise for the\nthird and fourth predictors. The fifth predictor, which has no effect\non $y$, potentially complicates matters because it is also strongly\ncorrelated with the other predictors. Despite this complication, our\nbasic goal remains the same: the Credible Sets inferred by SuSiE\nshould capture the true effects most of the time, while also\nminimizing \"false positive\" CSs that do not contain any true\neffects. (Further, each CS should, ideally, be as small as possible.)",
"_____no_output_____"
],
[
"Table below summarizes the results of these simulations:\nthe left-hand column gives a unique result (a combination of CSs), and\nthe right-hand column gives the number of times this unique result\noccurred among the 1,000 simulations. The CS combinations are ordered\nby the frequency of their occurrence in the simulations. We highlight\nin **bold** CSs that do not contain a true effect.\n\n| CSs | count |\n| :------------- | ----: |\n| (2), (3) | 551 |\n| (2), (3,4) | 212 |\n| (1,2), (3) | 176 |\n| (1,2), (3,4) | 38 |\n| (2), (3,4,5) | 9 |\n| **(1)**, (3,4) | 3 |\n| (2), **(4)** | 3 |\n| (1,2), (3,4,5) | 2 |\n| **(1)**, (3) | 1 |\n| (1,2), **(4)** | 1 |\n| (2), (3,5) | 1 |\n| (3), (1,2,5) | 1 |\n| (3), (1,2,3) | 1 |\n| (3,4), (1,2,4) | 1 |",
"_____no_output_____"
],
[
"In the majority (551) of the simulations, SuSiE precisely identiied\nthe true effect variables, and no others. In most other cases,\nSuSiE identified two CSs, each containing a correct effect variable, and\nwith one or more other variables included due to high correlation with\nthe true-effect variable. The referee asks specifically about how the\nadditional variable $x_5$ behaves in this example. In practice, $x_5$\nwas rarely included in a CS. In the few cases where $x_5$ *was*\nincluded in a CS, the results were consistent with the simulation\nsetting; $x_5$ was included more frequently with $x_3$ and/or $x_4$\n(12 times) rather than $x_2$ and/or $x_1$ (only once). In no\nsimulations did SuSiE form a large group that contains all five\npredictors.",
"_____no_output_____"
],
[
"This example actually highlights the benefits of SuSiE compared to\nalternative approaches (e.g., hierinf) that *first* cluster the\nvariables into groups based on the correlation structure, then test\nthe groups. As we pointed out in the manuscript, this alternative\napproach (first cluster variables into groups, then test groups) would\nwork well in the toy example in the paper, but in general it requires\n*ad hoc* decisions about how to cluster variables. In this more\ncomplex example raised by the referee, it is far from clear how to\ncluster the variables. SuSiE avoids this problem because there is\nno pre-clustering of variables; instead, the SuSiE CSs are computed\ndirectly from an (approximate) posterior distribution (which takes\ninto account how the variables $x$ are correlated with each other, as\nwell as their relationship with $y$).",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
d0b6828d6218788c650e77d990e28fd31d1b16f1 | 6,465 | ipynb | Jupyter Notebook | notebooks/01.4_data_inventory.ipynb | martinlarsalbert/datadriven-energieffektivisering-av-fartyg | 2fc65d611ff6d9497cb916b43477af9511912220 | [
"MIT"
] | 2 | 2021-04-19T09:04:51.000Z | 2021-11-14T19:41:15.000Z | notebooks/01.4_data_inventory.ipynb | martinlarsalbert/datadriven-energieffektivisering-av-fartyg | 2fc65d611ff6d9497cb916b43477af9511912220 | [
"MIT"
] | null | null | null | notebooks/01.4_data_inventory.ipynb | martinlarsalbert/datadriven-energieffektivisering-av-fartyg | 2fc65d611ff6d9497cb916b43477af9511912220 | [
"MIT"
] | null | null | null | 25.55336 | 111 | 0.536891 | [
[
[
"## Analyzing Hamlet",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import src.data\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport os\nfrom collections import OrderedDict\n\nfrom IPython.display import display\n\npd.options.display.max_rows = 999\npd.options.display.max_columns = 999\npd.set_option(\"display.max_columns\", None)",
"_____no_output_____"
],
[
"import itertools",
"_____no_output_____"
],
[
"file_names = {\n 'aurora':'2020-01-01-till-2021-02-24-aurora.csv',\n 'hamlet':'2020-01-01-till-2021-02-24-hamlet.csv',\n 'mercandia':'2020-01-01-till-2021-02-24-mercandia-iv.csv',\n 'tycho-brahe':'2020-01-01-till-2021-02-24-tycho-brahe.csv',\n}",
"_____no_output_____"
],
[
"dfs = OrderedDict()\n\nfor ship_name, file_name in file_names.items():\n \n file_path = os.path.join(src.data.path_ships,file_name)\n reader = pd.read_csv(file_path, chunksize=1000, iterator=True) # Loading a small part of the data\n dfs[ship_name] = next(reader)\n ",
"_____no_output_____"
],
[
"for ship_name, df in dfs.items():\n \n display(df.describe())\n ",
"_____no_output_____"
],
[
"file_path = os.path.join(src.data.path_ships,file_names['aurora']) \nreader = pd.read_csv(file_path, chunksize=1000000, iterator=True) # Loading a small part of the data\ndf_raw = next(reader)\ndf_raw.set_index('Tidpunkt [UTC]', inplace=True)\ndf_raw.index = pd.to_datetime(df_raw.index)",
"_____no_output_____"
],
[
"mask = df_raw['Fart över grund (kts)']>1\ndf = df_raw.loc[mask].copy()",
"_____no_output_____"
],
[
"df.hist(column='Kurs över grund (deg)', bins=1000)",
"_____no_output_____"
],
[
"mask = df_raw['Kurs över grund (deg)'] < 150\ndf_direction_1 = df.loc[mask]\ndf_direction_1.describe()",
"_____no_output_____"
],
[
"df_direction_1.plot(x='Longitud (deg)', y = 'Latitud (deg)', style='.', alpha=0.005)",
"_____no_output_____"
],
[
"deltas = []\nfor i in range(1,5):\n sin_key = 'Sin EM%i ()' % i\n cos_key = 'Cos EM%i ()' % i\n delta_key = 'delta_%i' % i\n deltas.append(delta_key)\n\n df_direction_1[delta_key] = np.arctan2(df_direction_1[sin_key],df_direction_1[cos_key])\n\ndf_plot = df_direction_1.loc['2020-01-01 01:00':'2020-01-01 02:00']\n \ndf_plot.plot(y=['Kurs över grund (deg)','Stävad kurs (deg)'],style='.')\ndf_plot.plot(y='Fart över grund (kts)',style='.')\n\ndf_plot.plot(y=deltas,style='.')\n\n",
"_____no_output_____"
],
[
"df_direction_1.head()",
"_____no_output_____"
],
[
"(df_direction_1['Sin EM1 ()']**2 + df_direction_1['Cos EM1 ()']**2).hist()",
"_____no_output_____"
],
[
"df_direction_1.columns",
"_____no_output_____"
],
[
"\ndescriptions = pd.Series(index = df_direction_1.columns.copy())\ndescriptions['Latitud (deg)'] = 'Latitud (deg) (WGS84?)'\ndescriptions['Longitud (deg)'] = 'Longitud (deg) (WGS84?)'\ndescriptions['Effekt DG Total (kW)'] = '?'\ndescriptions['Effekt EM Thruster Total (kW)'] = ''\ndescriptions['Sin EM1 ()'] = ''\ndescriptions['Sin EM2 ()'] = ''\ndescriptions['Sin EM3 ()'] = ''\ndescriptions['Sin EM4 ()'] = ''\ndescriptions['Cos EM1 ()'] = ''\ndescriptions['Cos EM2 ()'] = ''\ndescriptions['Cos EM3 ()'] = ''\ndescriptions['Cos EM4 ()'] = ''\ndescriptions['Fart över grund (kts)'] = 'GPS fart'\ndescriptions['Stävad kurs (deg)'] = 'Kompas kurs'\ndescriptions['Kurs över grund (deg)'] = 'GPS kurs'\ndescriptions['Effekt hotell Total (kW)'] = ''\ndescriptions['Effekt Consumption Total (kW)'] = ''\ndescriptions['Förbrukning GEN alla (kg/h)'] = '?'\ndescriptions['delta_1'] = 'Thruster angle 1'\ndescriptions['delta_2'] = 'Thruster angle 2'\ndescriptions['delta_3'] = 'Thruster angle 3'\ndescriptions['delta_4'] = 'Thruster angle 4'\n\ndf_numenclature = pd.DataFrame(descriptions, columns=['Description'])\ndf_numenclature",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b6af47e4b68f9d9a86dc29c4e340c685364dad | 6,745 | ipynb | Jupyter Notebook | sdk/jobs/pipelines/2a_train_mnist_with_tensorflow/train_mnist_with_tensorflow.ipynb | jplummer01/azureml-examples | 6a073d157f21060312941f71cfbcf25d0c541183 | [
"MIT"
] | null | null | null | sdk/jobs/pipelines/2a_train_mnist_with_tensorflow/train_mnist_with_tensorflow.ipynb | jplummer01/azureml-examples | 6a073d157f21060312941f71cfbcf25d0c541183 | [
"MIT"
] | null | null | null | sdk/jobs/pipelines/2a_train_mnist_with_tensorflow/train_mnist_with_tensorflow.ipynb | jplummer01/azureml-examples | 6a073d157f21060312941f71cfbcf25d0c541183 | [
"MIT"
] | null | null | null | 29.583333 | 341 | 0.611416 | [
[
[
"# Train mnist with Tensorflow\n\n**Requirements** - In order to benefit from this tutorial, you will need:\n- A basic understanding of Machine Learning\n- An Azure account with an active subscription - [Create an account for free](https://azure.microsoft.com/free/?WT.mc_id=A261C142F)\n- An Azure ML workspace with computer cluster - [Configure workspace](../../configuration.ipynb)\n- A python environment\n- Installed Azure Machine Learning Python SDK v2 - [install instructions](../../../README.md) - check the getting started section\n\n**Learning Objectives** - By the end of this tutorial, you should be able to:\n- Connect to your AML workspace from the Python SDK\n- Define different `CommandComponent` using YAML\n- Create `Pipeline` load these components from YAML\n\n**Motivations** - This notebook explains how to run a pipeline with distributed training component.",
"_____no_output_____"
],
[
"# 1. Connect to Azure Machine Learning Workspace\n\nThe [workspace](https://docs.microsoft.com/en-us/azure/machine-learning/concept-workspace) is the top-level resource for Azure Machine Learning, providing a centralized place to work with all the artifacts you create when you use Azure Machine Learning. In this section we will connect to the workspace in which the job will be run.\n\n## 1.1 Import the required libraries",
"_____no_output_____"
]
],
[
[
"# import required libraries\nfrom azure.identity import DefaultAzureCredential, InteractiveBrowserCredential\n\nfrom azure.ai.ml import MLClient\nfrom azure.ai.ml.dsl import pipeline\nfrom azure.ai.ml.entities import ResourceConfiguration",
"_____no_output_____"
]
],
[
[
"## 1.2 Configure credential\n\nWe are using `DefaultAzureCredential` to get access to workspace. \n`DefaultAzureCredential` should be capable of handling most Azure SDK authentication scenarios. \n\nReference for more available credentials if it does not work for you: [configure credential example](../../configuration.ipynb), [azure-identity reference doc](https://docs.microsoft.com/en-us/python/api/azure-identity/azure.identity?view=azure-python).",
"_____no_output_____"
]
],
[
[
"try:\n credential = DefaultAzureCredential()\n # Check if given credential can get token successfully.\n credential.get_token(\"https://management.azure.com/.default\")\nexcept Exception as ex:\n # Fall back to InteractiveBrowserCredential in case DefaultAzureCredential not work\n credential = InteractiveBrowserCredential()",
"_____no_output_____"
]
],
[
[
"## 1.3 Get a handle to the workspace\n\nWe use config file to connect to a workspace. The Azure ML workspace should be configured with computer cluster. [Check this notebook for configure a workspace](../../configuration.ipynb)",
"_____no_output_____"
]
],
[
[
"# Get a handle to workspace\nml_client = MLClient.from_config(credential=credential)\n\n# Retrieve an already attached Azure Machine Learning Compute.\ncluster_name = \"cpu-cluster\"\nprint(ml_client.compute.get(cluster_name))",
"_____no_output_____"
]
],
[
[
"# 2. Define command component\n\nWe defined sample component using `command_component` decorator in [component.py](src/component.py).",
"_____no_output_____"
]
],
[
[
"with open(\"src/component.py\") as fin:\n print(fin.read())",
"_____no_output_____"
],
[
"%load_ext autoreload\n%autoreload 2\n\nfrom src.component import train_tf\n\nhelp(train_tf)",
"_____no_output_____"
]
],
[
[
"# 3. Pipeline job\n## 3.1 Build pipeline",
"_____no_output_____"
]
],
[
[
"@pipeline()\ndef train_mnist_with_tensorflow():\n \"\"\"Train using TF component.\"\"\"\n tf_job = train_tf(epochs=1)\n tf_job.compute = \"cpu-cluster\"\n tf_job.resources = ResourceConfiguration(instance_count=2)\n tf_job.distribution.worker_count = 2\n tf_job.outputs.trained_model_output.mode = \"upload\"\n\n\n# create pipeline instance\npipeline_job = train_mnist_with_tensorflow()",
"_____no_output_____"
]
],
[
[
"## 3.2 Submit pipeline job",
"_____no_output_____"
]
],
[
[
"# submit job to workspace\npipeline_job = ml_client.jobs.create_or_update(\n pipeline_job, experiment_name=\"pipeline_samples\"\n)\npipeline_job",
"_____no_output_____"
],
[
"# Wait until the job completes\nml_client.jobs.stream(pipeline_job.name)",
"_____no_output_____"
]
],
[
[
"# Next Steps\nYou can see further examples of running a pipeline job [here](../)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d0b6d145f4ec6c6c9a23b87a44e98c2fbd42b58a | 2,179 | ipynb | Jupyter Notebook | load_write_json.ipynb | danielmlow/tutorials | 76147e179858f6a1de8541f43597854524bf450f | [
"Apache-2.0"
] | 1 | 2022-02-09T16:49:28.000Z | 2022-02-09T16:49:28.000Z | load_write_json.ipynb | danielmlow/tutorials | 76147e179858f6a1de8541f43597854524bf450f | [
"Apache-2.0"
] | null | null | null | load_write_json.ipynb | danielmlow/tutorials | 76147e179858f6a1de8541f43597854524bf450f | [
"Apache-2.0"
] | null | null | null | 25.045977 | 232 | 0.451124 | [
[
[
"<a href=\"https://colab.research.google.com/github/danielmlow/tutorials/blob/main/load_write_json.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import json\nimport datetime\n\ninput_dir = './'\noutput_dir = './'\n\nfilename = 'name'\n\ndata = {}\n\n# Save\nts = datetime.datetime.utcnow().strftime('%y-%m-%dT%H-%M-%S') # so you don't overwrite, and save timestamp\nwith open(output_dir+f'{filename}_{ts}.json', \"w\") as fp:\n json.dump(data, fp, indent=4) \n\n# Load \nwith open(input_dir+f'{filename}_{ts}.json', 'r') as json_file:\n data = json.load(json_file)\n\ndata",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
d0b6d1cd32d234311a958f00692277ce35bee9d0 | 28,029 | ipynb | Jupyter Notebook | Notebook/.ipynb_checkpoints/Morris_ranking_verification_GWP per Capita Indicator-checkpoint.ipynb | enayatmoallemi/Moallemi_et_al_SDG_SSP_Assessment | 552798d2ecee53b515abf2ca3cc72145e1b41113 | [
"BSD-3-Clause"
] | 1 | 2021-03-05T03:44:09.000Z | 2021-03-05T03:44:09.000Z | Notebook/.ipynb_checkpoints/Morris_ranking_verification_GWP per Capita Indicator-checkpoint.ipynb | enayatmoallemi/Moallemi_et_al_SDG_SSP_Assessment | 552798d2ecee53b515abf2ca3cc72145e1b41113 | [
"BSD-3-Clause"
] | null | null | null | Notebook/.ipynb_checkpoints/Morris_ranking_verification_GWP per Capita Indicator-checkpoint.ipynb | enayatmoallemi/Moallemi_et_al_SDG_SSP_Assessment | 552798d2ecee53b515abf2ca3cc72145e1b41113 | [
"BSD-3-Clause"
] | 1 | 2021-08-05T20:38:24.000Z | 2021-08-05T20:38:24.000Z | 42.792366 | 897 | 0.606515 | [
[
[
"import sys\nsys.path.append(r'C:\\Users\\moallemie\\EMAworkbench-master')\nsys.path.append(r'C:\\Users\\moallemie\\EM_analysis')",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom ema_workbench import load_results, ema_logging\nfrom ema_workbench.em_framework.salib_samplers import get_SALib_problem\nfrom SALib.analyze import morris",
"_____no_output_____"
],
[
"# Set up number of scenarios, outcome of interest, and number of parallel processors\n\nsc = 500 # Specify the number of scenarios where the convergence in the SA indices occures\nt = 2100\noutcome_var = 'GWP per Capita Indicator' # Specify the outcome of interest for SA ranking verification\nnprocess = 100",
"_____no_output_____"
]
],
[
[
"## Loading the model, uncertainities, and outcomes and generate experiments",
"_____no_output_____"
]
],
[
[
"# Here we only generate experiments for loading the necessary components. \n#The actual results will be loaded in the next cell.\n\n# Open Excel input data from the notebook directory before runnign the code in multi-processing.\n# Close the folder where the results will be saved in multi-processing.\n# This line must be at the beginning for multi processing. \nif __name__ == '__main__':\n \n \n ema_logging.log_to_stderr(ema_logging.INFO)\n \n #The model must be imoorted as .py file in parallel processing.\n from Model_init import vensimModel\n \n from ema_workbench import (TimeSeriesOutcome, \n perform_experiments,\n RealParameter, \n CategoricalParameter,\n ema_logging, \n save_results,\n load_results)\n\n directory = 'C:/Users/moallemie/EM_analysis/Model/'\n\n df_unc = pd.read_excel(directory+'ScenarioFramework.xlsx', sheet_name='Uncertainties')\n \n # 0.5/1.5 multiplication is added to previous Min/Max cells for parameters with Reference values 0 \n #or min/max manually set in the spreadsheet \n df_unc['Min'] = df_unc['Min'] + df_unc['Reference'] * 0.75\n df_unc['Max'] = df_unc['Max'] + df_unc['Reference'] * 1.25\n\n \n \n \n # From the Scenario Framework (all uncertainties), filter only those top 20 sensitive uncertainties under each outcome\n \n sa_dir='C:/Users/moallemie/EM_analysis/Data/'\n \n \n mu_df = pd.read_csv(sa_dir+\"MorrisIndices_{}_sc5000_t{}.csv\".format(outcome_var, t))\n mu_df.rename(columns={'Unnamed: 0': 'Uncertainty'}, inplace=True)\n mu_df.sort_values(by=['mu_star'], ascending=False, inplace=True)\n mu_df = mu_df.head(20)\n mu_unc = mu_df['Uncertainty']\n mu_unc_df = mu_unc.to_frame()\n \n \n # Remove the rest of insensitive uncertainties from the Scenario Framework and update df_unc\n keys = list(mu_unc_df.columns.values)\n i1 = df_unc.set_index(keys).index\n i2 = mu_unc_df.set_index(keys).index\n df_unc2 = df_unc[i1.isin(i2)]\n \n \n vensimModel.uncertainties = [RealParameter(row['Uncertainty'], row['Min'], row['Max']) for index, row in df_unc2.iterrows()]\n\n df_out = pd.read_excel(directory+'ScenarioFramework.xlsx', sheet_name='Outcomes')\n vensimModel.outcomes = [TimeSeriesOutcome(out) for out in df_out['Outcome']]\n\n\n \n from ema_workbench import MultiprocessingEvaluator\n from ema_workbench.em_framework.evaluators import (MC, LHS, FAST, FF, PFF, SOBOL, MORRIS)\n\n import time\n start = time.time()\n\n with MultiprocessingEvaluator(vensimModel, n_processes=nprocess) as evaluator:\n results = evaluator.perform_experiments(scenarios=sc, uncertainty_sampling=MORRIS)\n\n \n \n end = time.time()\n print(\"took {} seconds\".format(end-start))\n \n ",
"[MainProcess/INFO] using 64 bit vensim\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n"
],
[
"experiments, outcomes = results",
"_____no_output_____"
],
[
"r_dir = 'D:/moallemie/EM_analysis/Data/'\n\nsave_results(results, r_dir+\"SDG_experiments_ranking_verification_{}_sc{}.tar.gz\".format(outcome_var, sc))",
"[MainProcess/INFO] results saved successfully to D:\\moallemie\\EM_analysis\\Data\\SDG_experiments_ranking_verification_GWP per Capita Indicator_sc500.tar.gz\n"
]
],
[
[
"## Calculating SA (Morris) metrics",
"_____no_output_____"
]
],
[
[
"# Morris mu_star index calculation as a function of number of scenarios and time\n\n\ndef make_morris_df(scores, problem, outcome_var, sc, t):\n scores_filtered = {k:scores[k] for k in ['mu_star','mu_star_conf','mu','sigma']}\n Si_df = pd.DataFrame(scores_filtered, index=problem['names'])\n \n indices = Si_df[['mu_star','mu']]\n errors = Si_df[['mu_star_conf','sigma']]\n return indices, errors\n ",
"_____no_output_____"
],
[
"\nproblem = get_SALib_problem(vensimModel.uncertainties)\nX = experiments.iloc[:, :-3].values\nY = outcomes[outcome_var][:,-1]\nscores = morris.analyze(problem, X, Y, print_to_console=False)\ninds, errs = make_morris_df(scores, problem, outcome_var, sc, t)\n",
"_____no_output_____"
]
],
[
[
"## Where to draw the line between important and not important?",
"_____no_output_____"
]
],
[
[
"'''\nModifed from Waterprogramming blog by Antonia Hadgimichael: https://github.com/antonia-had/SA_verification\n\nThe idea is that we create 2 additiopnal Sets (current SA samples are Set 1).\n\nWe can create a Set 2, using only the T most important factors from our Set 1 sample, \nand fixing all other factors to their default values.\n\nWe can also create a Set 3, now fixing the T most important factors to defaults \nand using the sampled values of all other factors from Set 1.\n\nIf we classified our important and unimportant factors correctly, \nthen the correlation coefficient between the model outputs of Set 2 and Set 1 should approximate 1 \n(since we’re fixing all factors that don’t matter), \nand the correlation coefficient between outputs from Set 3 and Set 1 should approximate 0 \n(since the factors we sampled are inconsequential to the output).\n\n'''",
"_____no_output_____"
],
[
"# Sort factors by importance\ninds_mu = inds['mu_star'].reindex(df_unc2['Uncertainty']).values\nfactors_sorted = np.argsort(inds_mu)[::-1]\n\n# Set up DataFrame of default values to use for experiment\nnsamples = len(experiments.index)\n\ndefaultvalues = df_unc2['Reference'].values\nX_defaults = np.tile(defaultvalues,(nsamples, 1))\n\n\n# Create Set 1 from experiments\nexp_T = experiments.drop(['scenario', 'policy', 'model'], axis=1).T.reindex(df_unc2['Uncertainty'])\nexp_ordered = exp_T.T\nX_Set1 = exp_ordered.values\n\n# Create initial Sets 2 and 3\nX_Set2 = np.copy(X_defaults)\nX_Set3 = np.copy(X_Set1)",
"_____no_output_____"
],
[
"# Define a function to convert your Set 2 and Set 3 into experiments structure in the EMA Workbench\n\ndef SA_experiments_to_scenarios(experiments, model=None):\n '''\n\n \"Slighlty modifed from the EMA Workbench\"\n \n This function transform a structured experiments array into a list\n of Scenarios.\n\n If model is provided, the uncertainties of the model are used.\n Otherwise, it is assumed that all non-default columns are\n uncertainties.\n\n Parameters\n ----------\n experiments : numpy structured array\n a structured array containing experiments\n model : ModelInstance, optional\n\n Returns\n -------\n a list of Scenarios\n\n '''\n from ema_workbench import Scenario\n \n # get the names of the uncertainties\n uncertainties = [u.name for u in model.uncertainties]\n\n # make list of of tuples of tuples\n cases = []\n cache = set()\n for i in range(experiments.shape[0]):\n case = {}\n case_tuple = []\n for uncertainty in uncertainties:\n entry = experiments[uncertainty][i]\n case[uncertainty] = entry\n case_tuple.append(entry)\n\n case_tuple = tuple(case_tuple)\n cases.append(case)\n cache.add((case_tuple))\n\n scenarios = [Scenario(**entry) for entry in cases]\n\n return scenarios",
"_____no_output_____"
],
[
"# Run the models for the top n factors in Set 2 and Set 3 and generate correlation figures\n\nif __name__ == '__main__':\n \n ema_logging.log_to_stderr(ema_logging.INFO)\n \n #The model must be imoorted as .py file in parallel processing.\n from Model_init import vensimModel\n \n from ema_workbench import (TimeSeriesOutcome, \n perform_experiments,\n RealParameter, \n CategoricalParameter,\n ema_logging, \n save_results,\n load_results)\n \n vensimModel.outcomes = [TimeSeriesOutcome(outcome_var)]\n\n from ema_workbench import MultiprocessingEvaluator\n \n coefficient_S1_S3 = 0.99\n \n for f in range(1, len(factors_sorted)+1):\n ntopfactors = f\n \n if coefficient_S1_S3 >= 0.1:\n\n for i in range(ntopfactors): #Loop through all important factors\n X_Set2[:,factors_sorted[i]] = X_Set1[:,factors_sorted[i]] #Fix use samples for important\n X_Set3[:,factors_sorted[i]] = X_defaults[:,factors_sorted[i]] #Fix important to defaults\n\n X_Set2_exp = pd.DataFrame(data=X_Set2, columns=df_unc2['Uncertainty'].tolist())\n X_Set3_exp = pd.DataFrame(data=X_Set3, columns=df_unc2['Uncertainty'].tolist())\n\n scenarios_Set2 = SA_experiments_to_scenarios(X_Set2_exp, model=vensimModel)\n scenarios_Set3 = SA_experiments_to_scenarios(X_Set3_exp, model=vensimModel)\n\n #experiments_Set2, outcomes_Set2 = perform_experiments(vensimModel, scenarios_Set2)\n #experiments_Set3, outcomes_Set3 = perform_experiments(vensimModel, scenarios_Set3)\n with MultiprocessingEvaluator(vensimModel, n_processes=nprocess) as evaluator:\n experiments_Set2, outcomes_Set2 = evaluator.perform_experiments(scenarios=scenarios_Set2)\n experiments_Set3, outcomes_Set3 = evaluator.perform_experiments(scenarios=scenarios_Set3)\n\n\n # Calculate coefficients of correlation\n data_Set1 = Y\n data_Set2 = outcomes_Set2[outcome_var][:,-1]\n data_Set3 = outcomes_Set3[outcome_var][:,-1]\n\n coefficient_S1_S2 = np.corrcoef(data_Set1,data_Set2)[0][1]\n coefficient_S1_S3 = np.corrcoef(data_Set1,data_Set3)[0][1]\n\n # Plot outputs and correlation\n fig = plt.figure(figsize=(14,7))\n ax1 = fig.add_subplot(1,2,1)\n ax1.plot(data_Set1,data_Set1, color='#39566E')\n ax1.scatter(data_Set1,data_Set2, color='#8DCCFC')\n ax1.set_xlabel(\"Set 1\",fontsize=14)\n ax1.set_ylabel(\"Set 2\",fontsize=14)\n ax1.tick_params(axis='both', which='major', labelsize=10)\n ax1.set_title('Set 1 vs Set 2 - ' + str(f) + ' top factors',fontsize=15)\n ax1.text(0.05,0.95,'R= '+\"{0:.3f}\".format(coefficient_S1_S2),transform = ax1.transAxes,fontsize=16)\n ax2 = fig.add_subplot(1,2,2)\n ax2.plot(data_Set1,data_Set1, color='#39566E')\n ax2.scatter(data_Set1,data_Set3, color='#FFE0D5')\n ax2.set_xlabel(\"Set 1\",fontsize=14)\n ax2.set_ylabel(\"Set 3\",fontsize=14)\n ax2.tick_params(axis='both', which='major', labelsize=10)\n ax2.set_title('Set 1 vs Set 3 - ' + str(f) + ' top factors',fontsize=15)\n ax2.text(0.05,0.95,'R= '+\"{0:.3f}\".format(coefficient_S1_S3),transform = ax2.transAxes,fontsize=16)\n plt.savefig('{}/{}_{}_topfactors.png'.format(r'C:/Users/moallemie/EM_analysis/Fig/sa_verification', outcome_var, str(f)))\n plt.close()",
"[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n[MainProcess/INFO] pool started\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] performing 10500 scenarios * 1 policies * 1 model(s) = 10500 experiments\n[MainProcess/INFO] 1050 cases completed\n[MainProcess/INFO] 2100 cases completed\n[MainProcess/INFO] 3150 cases completed\n[MainProcess/INFO] 4200 cases completed\n[MainProcess/INFO] 5250 cases completed\n[MainProcess/INFO] 6300 cases completed\n[MainProcess/INFO] 7350 cases completed\n[MainProcess/INFO] 8400 cases completed\n[MainProcess/INFO] 9450 cases completed\n[MainProcess/INFO] 10500 cases completed\n[MainProcess/INFO] experiments finished\n[MainProcess/INFO] terminating pool\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0b6d6f399c29408599e8f871eb33d8a945c0af3 | 53,492 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Apriori_Algo-checkpoint.ipynb | MichaelWoo-git/Apriori_Algorithm_from_Scratch | b13416bb6343f4b4cdd4e5ca470301b3cd78ce4e | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Apriori_Algo-checkpoint.ipynb | MichaelWoo-git/Apriori_Algorithm_from_Scratch | b13416bb6343f4b4cdd4e5ca470301b3cd78ce4e | [
"MIT"
] | null | null | null | .ipynb_checkpoints/Apriori_Algo-checkpoint.ipynb | MichaelWoo-git/Apriori_Algorithm_from_Scratch | b13416bb6343f4b4cdd4e5ca470301b3cd78ce4e | [
"MIT"
] | null | null | null | 35.168968 | 152 | 0.406734 | [
[
[
"##### Imports",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport os\nimport time\nfrom itertools import permutations, combinations\nfrom IPython.display import display",
"_____no_output_____"
]
],
[
[
"##### Prompts to choose which store you want ",
"_____no_output_____"
]
],
[
[
"print(\"Welcome to Apriori 2.0!\")\nstore_num = input(\"Please select your store \\n 1. Amazon \\n 2. Nike \\n 3. Best Buy \\n 4. K-Mart \\n 5. Walmart\\n\")\nprint(store_num)\nsupport_percent = input(\"Please enter the percentage of Support you want?\\n\")\nprint(support_percent)\nconfidence_percent = input(\"Please enter the percentage of Confidence you want?\\n\")\nprint(confidence_percent)",
"Welcome to Apriori 2.0!\nPlease select your store \n 1. Amazon \n 2. Nike \n 3. Best Buy \n 4. K-Mart \n 5. Walmart\n1\n1\nPlease enter the percentage of Support you want?\n40\n40\nPlease enter the percentage of Confidence you want?\n40\n40\n"
]
],
[
[
"##### These are my dictionaries to choose which store to get based in Key-Value Pairs",
"_____no_output_____"
]
],
[
[
"def number_to_store(store_number):\n switcher = {\n 1: \"data/amazon_transactions.csv\",\n 2: \"data/nike_transaction.csv\",\n 3: \"data/best_buy_transaction.csv\",\n 4: \"data/k_mart_transaction.csv\",\n 5: \"data/walmart_transaction.csv\"\n }\n return switcher.get(store_number)\n\n\ndef number_to_item_list_of_store(store_number):\n switcher_dict = {\n 1: \"data/amazon_item_names.csv\",\n 2: \"data/nike_item_names.csv\",\n 3: \"data/best_buy_item_names.csv\",\n 4: \"data/k_mart_item_names.csv\",\n 5: \"data/walmart_item_names.csv\"\n }\n return switcher_dict.get(store_number)\n\ndef ns(store_number):\n switcher_store = {\n 1: \"Amazon\",\n 2: \"Nike\",\n 3: \"Best Buy\",\n 4: \"K-Mart\",\n 5: \"Walmart\"\n }\n return switcher_store.get(store_number)",
"_____no_output_____"
]
],
[
[
"##### We first have to read in the csv files and make sure that the inputs received from the user are valid",
"_____no_output_____"
]
],
[
[
"def a_priori_read(item_list, transaction, support_percentage, confidence_percentage):\n # Create two different functions one that is solo for read in the file data and the other that is algorithmic with the data\n if support_percentage > 100 or confidence_percentage > 100 or support_percentage < 0 or confidence_percentage < 0:\n print(\"Support Percent or Confidence Percent is Invalid. \\n Enter a valid number between 0 and 100.\\n\")\n print(\"Restarting Apriori 2.0.....\\n\")\n time.sleep(2)\n os.system(\"python Aprior_Algo\")\n if support_percentage >= 0 and support_percentage <= 100 and confidence_percentage >= 0 and confidence_percentage <= 100:\n df_item_list = pd.read_csv(item_list)\n df_transactions = pd.read_csv(transaction)\n print(df_transactions.head())\n print(df_item_list.head())\n trans = np.array(df_transactions[\"transaction\"])\n items_names = np.array(df_item_list[\"item_name\"])\n k_value = 1\n return items_names, trans, support_percentage, confidence_percentage, k_value",
"_____no_output_____"
]
],
[
[
"##### The first go around of the Apriori Algorithm we find the items that are most frequent when K=1\n##### This is so that we can find the most frequent items given the transactions",
"_____no_output_____"
]
],
[
[
"def ap_1(items_names, trans, support_percentage, confidence_percentage, k_value):\n counter = np.zeros(len(items_names), dtype=int)\n for i in trans:\n i = list((map(str.strip, i.split(','))))\n s1 = set(i)\n nums = 0\n for x in items_names:\n s2 = set()\n s2.add(x)\n if s2.issubset(s1):\n counter[nums] += 1\n nums += 1\n counter = list(map(lambda x: int((x / len(trans)) * 100), counter))\n df3 = pd.DataFrame({\"item_name\": items_names, \"support\": counter,\"k_val\" : np.full(len(items_names),k_value)})\n rslt_df = df3[df3['support'] >= support_percentage]\n print(\"When K = \" + str(k_value))\n print(rslt_df)\n items = np.array(rslt_df[\"item_name\"])\n support_count = np.array(rslt_df[\"support\"])\n k_value += 1\n return items, support_count, k_value, rslt_df",
"_____no_output_____"
]
],
[
[
"##### Then we use this function below to find item sets that are most frequent when K > 1",
"_____no_output_____"
]
],
[
[
"def ap_2(item_comb, k_value, trans, support_percentage):\n boo = True\n comb = combinations(item_comb, k_value)\n comb = list(comb)\n counter = np.zeros(len(comb), dtype=int)\n if k_value > 1:\n for i in trans:\n i = list((map(str.strip, i.split(','))))\n s1 = set(i)\n nums = 0\n for x in comb:\n s2 = set()\n x = np.asarray(x)\n for q in x:\n s2.add(q)\n if s2.issubset(s1):\n counter[nums] += 1\n nums += 1\n counter = list(map(lambda x: int((x / len(trans)) * 100), counter))\n df3 = pd.DataFrame({\"item_name\": comb, \"support\": counter,\"k_val\":np.full(len(comb),k_value)})\n \n #Making sure that user parameters are met for support\n rslt_df = df3[df3['support'] >= support_percentage]\n print(\"When K = \" + str(k_value))\n print(rslt_df)\n items = np.array(rslt_df[\"item_name\"])\n supp = np.array(rslt_df[\"support\"])\n if len(items) == 0:\n boo = False\n return rslt_df, boo\n return rslt_df, boo",
"_____no_output_____"
]
],
[
[
"##### Calls of functions and variable saving",
"_____no_output_____"
]
],
[
[
"frames = []\nitems_names, trans, support_percent, confidence_percent, k_value = a_priori_read(\n str(number_to_item_list_of_store(int(store_num))), str(number_to_store(int(store_num))),\n int(support_percent), int(confidence_percent))\n\nitems, supp, k_value, df = ap_1(items_names, trans, support_percent, confidence_percent, k_value)\nframes.append(df)\nboo = True",
" transaction_id \\\n0 1 \n1 2 \n2 3 \n3 4 \n4 5 \n\n transaction \n0 A Beginner’s Guide, Java: The Complete Reference, Java For Dummies, Android Programming: The Big Nerd Ranch \n1 A Beginner’s Guide, Java: The Complete Reference, Java For Dummies \n2 A Beginner’s Guide, Java: The Complete Reference, Java For Dummies, Android Programming: The Big Nerd Ranch, Head First Java 2nd Edition \n3 Android Programming: The Big Nerd Ranch, Head First Java 2nd Edition , Beginning Programming with Java, \n4 Android Programming: The Big Nerd Ranch, Beginning Programming with Java, Java 8 Pocket Guide \n item_number item_name\n0 1 A Beginner’s Guide \n1 2 Java: The Complete Reference \n2 3 Java For Dummies \n3 4 Android Programming: The Big Nerd Ranch\n4 5 Head First Java 2nd Edition \nWhen K = 1\n item_name support k_val\n0 A Beginner’s Guide 55 1 \n1 Java: The Complete Reference 50 1 \n2 Java For Dummies 65 1 \n3 Android Programming: The Big Nerd Ranch 65 1 \n4 Head First Java 2nd Edition 40 1 \n"
]
],
[
[
"##### Increasing K by 1 until we can longer support the support value",
"_____no_output_____"
]
],
[
[
"while boo:\n df_1, boo = ap_2(items, k_value, trans, support_percent)\n frames.append(df_1)\n k_value += 1",
"When K = 2\n item_name support k_val\n0 (A Beginner’s Guide, Java: The Complete Reference) 45 2 \n1 (A Beginner’s Guide, Java For Dummies) 45 2 \n4 (Java: The Complete Reference, Java For Dummies) 50 2 \n7 (Java For Dummies, Android Programming: The Big Nerd Ranch) 45 2 \nWhen K = 3\n item_name \\\n0 (A Beginner’s Guide, Java: The Complete Reference, Java For Dummies) \n\n support k_val \n0 45 3 \nWhen K = 4\nEmpty DataFrame\nColumns: [item_name, support, k_val]\nIndex: []\n"
]
],
[
[
"##### Combine the dataframes we have from when we increase K",
"_____no_output_____"
]
],
[
[
"print(\"results of item-sets that meet support are below\")\ndisplay(pd.concat(frames))\ndf_supp = pd.concat(frames)\n# df_supp.head()",
"results of item-sets that meet support are below\n"
]
],
[
[
"##### Reset the index just to organize it and the results after we find the most frequent sets in the list of transactions",
"_____no_output_____"
]
],
[
[
"df_supp = df_supp.reset_index().drop('index',axis=1)\ndf_supp",
"_____no_output_____"
]
],
[
[
"##### This is the FUNCTION that genrerates the Associations (Permutations) and calculating the Confidence of the item sets ",
"_____no_output_____"
]
],
[
[
"def confidence(val):\n \n #Since we already have our support for our items what we need to worry about is the confidence levels\n #item_set before the arrow\n \n df_before = df_supp.loc[df_supp['k_val'] == val] \n stuff_name_before = np.array(df_before[\"item_name\"])\n support_arr_before = np.array(df_before['support'])\n \n #item_set of the overall set\n df_overall = df_supp.loc[df_supp['k_val'] == val+1] \n df_ov = np.array(df_overall[\"item_name\"])\n suppport_ov = np.array(df_overall['support'])\n \n #variables to save\n sup_ov = list()\n sup_sing = list()\n perm_item = list()\n \n #When the item set is k =1 and the comparison is k = 2\n if val == 1:\n for i_set in df_ov:\n temp_list = list(df_ov)\n #I want to select the support of that overall set\n ov_sup = suppport_ov[temp_list.index(i_set)]\n temp = set()\n #This is where we generate our permutations \n for indiv_item in i_set:\n temp.add(indiv_item)\n perm = permutations(temp)\n perm_lst = list(perm)\n # for each permutation in the perm_list\n for perm_item_set in perm_lst:\n perm_item.append(perm_item_set)\n sup_ov.append(ov_sup)\n sup_sing.append(int(support_arr_before[np.where(stuff_name_before == perm_item_set[0])]))\n \n #When the item set is k > 1 and the comparison is k += k + 1\n if val > 1: \n for i_set in df_ov:\n temp_list = list(df_ov)\n ov_sup = suppport_ov[temp_list.index(i_set)]\n temp = set()\n for indiv_item in i_set:\n temp.add(indiv_item)\n perm = permutations(temp)\n perm_lst = list(perm)\n for perm_item_set in perm_lst:\n try:\n temp_set = []\n for dex in range(0,val):\n temp_set.append(perm_item_set[dex])\n item_set_before = tuple(temp_set)\n tp_lst = list(stuff_name_before)\n ss = support_arr_before[tp_lst.index(item_set_before)]\n sup_ov.append(ov_sup)\n sup_sing.append(ss)\n perm_item.append(perm_item_set)\n except:\n# print(\"itemset below does not exist...\")\n# print(y)\n sup_ov.append(ov_sup)\n sup_sing.append(0)\n perm_item.append(perm_item_set)\n \n df_main = pd.DataFrame({\"association\":perm_item,\"support_ov\":sup_ov,\"support_sing\":sup_sing})\n df_main = df_main.assign(confidence = lambda x:round(((x.support_ov/x.support_sing)*100),0))\n return df_main",
"_____no_output_____"
]
],
[
[
"#### Finding the max k value in the given set",
"_____no_output_____"
]
],
[
[
"try:\n max(df_supp[\"k_val\"])\nexcept:\n print(\"No max was found...\")",
"_____no_output_____"
]
],
[
[
"#### This is where I iteratively call the confidence() function",
"_____no_output_____"
]
],
[
[
"df_frames = []\ntry:\n if len(df_supp[\"k_val\"]) != 0 : \n for lp in range(1,max(df_supp[\"k_val\"])+1):\n #print(lp)\n df_0 = confidence(lp)\n df_0 = df_0[df_0.support_sing != 0]\n df_frames.append(df_0)\n df_associations = pd.concat(df_frames)\n display(df_associations.head())\nexcept:\n print(\"No items or transactions meet the user requirements!\")",
"_____no_output_____"
]
],
[
[
"###### Concat the Dataframes",
"_____no_output_____"
]
],
[
[
"try:\n df_associations = pd.concat(df_frames)\n display(df_associations)\nexcept:\n print(\"No items or transactions meet the user requirements!\")",
"_____no_output_____"
]
],
[
[
"##### Making sure that user parameters are met for confidence",
"_____no_output_____"
]
],
[
[
"try:\n df_associations = df_associations[df_associations['confidence'] >= confidence_percent]\n display(df_associations)\nexcept:\n print(\"No items or transactions meet the user requirements!\")",
"_____no_output_____"
]
],
[
[
"##### Formatting the Dataframe Final",
"_____no_output_____"
]
],
[
[
"try:\n df_final = df_associations.reset_index().drop(['index','support_sing'],axis=1)\n df_final.columns = [\"Association\",\"Support\",\"Confidence\"]\nexcept:\n print(\"No items or transactions meet the user requirements!\")",
"_____no_output_____"
]
],
[
[
"#### Final Associations ",
"_____no_output_____"
]
],
[
[
"try:\n print(\"Store Name: \"+ str(ns(int(store_num))))\n print(\"\\nFinal Associations that meet the user standards....\")\n print(\"Support: \" + str(support_percent) + \"%\" + \"\\t\" + \"Confidence: \" + str(confidence_percent) + '%')\n #this will display the max column width so we can see the associations involved....\n pd.set_option('display.max_colwidth', 0)\n display(df_final)\nexcept:\n print(\"\\nNo Associations were generated based on the parameters set!\")",
"Store Name: Amazon\n\nFinal Associations that meet the user standards....\nSupport: 40%\tConfidence: 40%\n"
],
[
"import re\nsamp = np.array(df_final.Association)\nwith_arrow = list()\nfor i in samp:\n left = str(i[0:(len(i)-number_of_assoications)])\n left = re.sub('[\\(\\)\\{\\}<>\\'''\\,]', '', left)\n right = i[(len(i)-number_of_assoications)]\n rslt = left + \" ==> \"+right\n with_arrow.append(rslt)\ndf_final.Association = with_arrow",
"_____no_output_____"
],
[
"df_final",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0b6d70ebb0dd54ad68444f67ba427142ffdb99a | 579,986 | ipynb | Jupyter Notebook | sphinx/pytorch-intro/source/neural-transfer.ipynb | oneoffcoder/books | 84619477294a3e37e0d7538adf819113c9e8dcb8 | [
"CC-BY-4.0"
] | 26 | 2020-05-05T08:07:43.000Z | 2022-02-12T03:28:15.000Z | sphinx/pytorch-intro/source/neural-transfer.ipynb | oneoffcoder/books | 84619477294a3e37e0d7538adf819113c9e8dcb8 | [
"CC-BY-4.0"
] | 19 | 2021-03-10T00:33:51.000Z | 2022-03-02T13:04:32.000Z | sphinx/pytorch-intro/source/neural-transfer.ipynb | oneoffcoder/books | 84619477294a3e37e0d7538adf819113c9e8dcb8 | [
"CC-BY-4.0"
] | 2 | 2022-01-09T16:48:21.000Z | 2022-02-19T17:06:50.000Z | 973.130872 | 192,284 | 0.953399 | [
[
[
"# Neural Transfer",
"_____no_output_____"
],
[
"## Input images",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\n\nfrom PIL import Image\nimport matplotlib.pyplot as plt\n\nimport torchvision.transforms as transforms\nimport torchvision.models as models\n\nimport copy\n\nnp.random.seed(37)\ntorch.manual_seed(37)\ntorch.backends.cudnn.deterministic = True\ntorch.backends.cudnn.benchmark = False\n\ndef get_device():\n return torch.device('cuda' if torch.cuda.is_available() else 'cpu')\n\ndef get_image_size():\n imsize = 512 if torch.cuda.is_available() else 128\n return imsize\n\ndef get_loader():\n image_size = get_image_size()\n loader = transforms.Compose([\n transforms.Resize((image_size, image_size)),\n transforms.ToTensor()]) \n return loader\n\ndef get_unloader():\n unloader = transforms.ToPILImage()\n return unloader\n\ndef image_loader(image_name):\n device = get_device()\n image = Image.open(image_name)\n # fake batch dimension required to fit network's input dimensions\n loader = get_loader()\n image = loader(image).unsqueeze(0)\n return image.to(device, torch.float)\n\ndef imshow(tensor, title=None):\n image = tensor.cpu().clone() # we clone the tensor to not do changes on it\n image = image.squeeze(0) # remove the fake batch dimension\n unloader = get_unloader()\n image = unloader(image)\n plt.imshow(image)\n if title is not None:\n plt.title(title)\n plt.pause(0.001) \n\nstyle_img = image_loader(\"./styles/picasso-01.jpg\")\ncontent_img = image_loader(\"./styles/dancing.jpg\")\ninput_img = content_img.clone()\n\nassert style_img.size() == content_img.size(), \\\n f'size mismatch, style {style_img.size()}, content {content_img.size()}'\n\nplt.ion()\n\nplt.figure()\nimshow(input_img, title='Input Image')\n\nplt.figure()\nimshow(style_img, title='Style Image')\n\nplt.figure()\nimshow(content_img, title='Content Image')",
"_____no_output_____"
]
],
[
[
"## Loss functions",
"_____no_output_____"
],
[
"### Content loss",
"_____no_output_____"
]
],
[
[
"class ContentLoss(nn.Module):\n\n def __init__(self, target,):\n super(ContentLoss, self).__init__()\n # we 'detach' the target content from the tree used\n # to dynamically compute the gradient: this is a stated value,\n # not a variable. Otherwise the forward method of the criterion\n # will throw an error.\n self.target = target.detach()\n\n def forward(self, input):\n self.loss = F.mse_loss(input, self.target)\n return input",
"_____no_output_____"
]
],
[
[
"### Style loss",
"_____no_output_____"
]
],
[
[
"def gram_matrix(input):\n a, b, c, d = input.size() # a=batch size(=1)\n # b=number of feature maps\n # (c,d)=dimensions of a f. map (N=c*d)\n\n features = input.view(a * b, c * d) # resise F_XL into \\hat F_XL\n\n G = torch.mm(features, features.t()) # compute the gram product\n\n # we 'normalize' the values of the gram matrix\n # by dividing by the number of element in each feature maps.\n return G.div(a * b * c * d)\n\nclass StyleLoss(nn.Module):\n\n def __init__(self, target_feature):\n super(StyleLoss, self).__init__()\n self.target = gram_matrix(target_feature).detach()\n\n def forward(self, input):\n G = gram_matrix(input)\n self.loss = F.mse_loss(G, self.target)\n return input",
"_____no_output_____"
]
],
[
[
"## Model",
"_____no_output_____"
]
],
[
[
"device = get_device()\n\ncnn = models.vgg19(pretrained=True).features.to(device).eval()",
"_____no_output_____"
]
],
[
[
"## Normalization",
"_____no_output_____"
]
],
[
[
"class Normalization(nn.Module):\n def __init__(self, mean, std):\n super(Normalization, self).__init__()\n # .view the mean and std to make them [C x 1 x 1] so that they can\n # directly work with image Tensor of shape [B x C x H x W].\n # B is batch size. C is number of channels. H is height and W is width.\n self.mean = torch.tensor(mean).view(-1, 1, 1)\n self.std = torch.tensor(std).view(-1, 1, 1)\n\n def forward(self, img):\n # normalize img\n return (img - self.mean) / self.std\n \ncnn_normalization_mean = torch.tensor([0.485, 0.456, 0.406]).to(device)\ncnn_normalization_std = torch.tensor([0.229, 0.224, 0.225]).to(device)",
"_____no_output_____"
]
],
[
[
"## Loss",
"_____no_output_____"
]
],
[
[
"content_layers_default = ['conv_4']\nstyle_layers_default = ['conv_1', 'conv_2', 'conv_3', 'conv_4', 'conv_5']\n\ndef get_style_model_and_losses(cnn, normalization_mean, normalization_std,\n style_img, content_img,\n content_layers=content_layers_default,\n style_layers=style_layers_default):\n cnn = copy.deepcopy(cnn)\n\n # normalization module\n normalization = Normalization(normalization_mean, normalization_std).to(device)\n\n # just in order to have an iterable access to or list of content/syle\n # losses\n content_losses = []\n style_losses = []\n\n # assuming that cnn is a nn.Sequential, so we make a new nn.Sequential\n # to put in modules that are supposed to be activated sequentially\n model = nn.Sequential(normalization)\n\n i = 0 # increment every time we see a conv\n for layer in cnn.children():\n if isinstance(layer, nn.Conv2d):\n i += 1\n name = 'conv_{}'.format(i)\n elif isinstance(layer, nn.ReLU):\n name = 'relu_{}'.format(i)\n # The in-place version doesn't play very nicely with the ContentLoss\n # and StyleLoss we insert below. So we replace with out-of-place\n # ones here.\n layer = nn.ReLU(inplace=False)\n elif isinstance(layer, nn.MaxPool2d):\n name = 'pool_{}'.format(i)\n elif isinstance(layer, nn.BatchNorm2d):\n name = 'bn_{}'.format(i)\n else:\n raise RuntimeError('Unrecognized layer: {}'.format(layer.__class__.__name__))\n\n model.add_module(name, layer)\n\n if name in content_layers:\n # add content loss:\n target = model(content_img).detach()\n content_loss = ContentLoss(target)\n model.add_module(\"content_loss_{}\".format(i), content_loss)\n content_losses.append(content_loss)\n\n if name in style_layers:\n # add style loss:\n target_feature = model(style_img).detach()\n style_loss = StyleLoss(target_feature)\n model.add_module(\"style_loss_{}\".format(i), style_loss)\n style_losses.append(style_loss)\n\n # now we trim off the layers after the last content and style losses\n for i in range(len(model) - 1, -1, -1):\n if isinstance(model[i], ContentLoss) or isinstance(model[i], StyleLoss):\n break\n\n model = model[:(i + 1)]\n\n return model, style_losses, content_losses",
"_____no_output_____"
]
],
[
[
"## Optimizer",
"_____no_output_____"
]
],
[
[
"def get_input_optimizer(input_img):\n # this line to show that input is a parameter that requires a gradient\n optimizer = optim.LBFGS([input_img.requires_grad_()])\n return optimizer",
"_____no_output_____"
]
],
[
[
"## Transfer",
"_____no_output_____"
]
],
[
[
"import warnings\nfrom collections import namedtuple\n\nRESULTS = namedtuple('RESULTS', 'run style content')\nresults = []\n\ndef run_style_transfer(cnn, normalization_mean, normalization_std,\n content_img, style_img, input_img, num_steps=600,\n style_weight=1000000, content_weight=1):\n model, style_losses, content_losses = get_style_model_and_losses(cnn,\n normalization_mean, normalization_std, style_img, content_img)\n optimizer = get_input_optimizer(input_img)\n\n run = [0]\n while run[0] <= num_steps:\n\n def closure():\n # correct the values of updated input image\n input_img.data.clamp_(0, 1)\n\n optimizer.zero_grad()\n model(input_img)\n style_score = 0\n content_score = 0\n\n for sl in style_losses:\n style_score += sl.loss\n for cl in content_losses:\n content_score += cl.loss\n\n style_score *= style_weight\n content_score *= content_weight\n\n loss = style_score + content_score\n loss.backward()\n\n run[0] += 1\n results.append(RESULTS(run[0], style_score.item(), content_score.item()))\n \n if run[0] % 10 == 0:\n s_score = style_score.item()\n c_score = content_score.item()\n \n print(f'[{run[0]}/{num_steps}] Style Loss {s_score:.4f}, Content Loss {c_score}') \n return style_score + content_score\n\n optimizer.step(closure)\n\n # a last correction...\n input_img.data.clamp_(0, 1)\n\n return input_img\n\nwith warnings.catch_warnings():\n warnings.simplefilter('ignore')\n output = run_style_transfer(cnn, cnn_normalization_mean, cnn_normalization_std,\n content_img, style_img, input_img)",
"[10/600] Style Loss 5683.9438, Content Loss 28.085737228393555\n[20/600] Style Loss 1513.7101, Content Loss 32.92472839355469\n[30/600] Style Loss 839.8792, Content Loss 33.72367477416992\n[40/600] Style Loss 495.5242, Content Loss 33.512489318847656\n[50/600] Style Loss 356.6638, Content Loss 33.42991256713867\n[60/600] Style Loss 269.5034, Content Loss 33.4761962890625\n[70/600] Style Loss 216.9316, Content Loss 33.47385787963867\n[80/600] Style Loss 163.2883, Content Loss 33.37232208251953\n[90/600] Style Loss 135.9556, Content Loss 33.303672790527344\n[100/600] Style Loss 109.1580, Content Loss 33.22367477416992\n[110/600] Style Loss 91.8495, Content Loss 33.01668930053711\n[120/600] Style Loss 79.5773, Content Loss 32.93888854980469\n[130/600] Style Loss 68.3634, Content Loss 32.801029205322266\n[140/600] Style Loss 57.8147, Content Loss 32.63578796386719\n[150/600] Style Loss 49.9333, Content Loss 32.42307662963867\n[160/600] Style Loss 44.3781, Content Loss 32.22199630737305\n[170/600] Style Loss 38.0064, Content Loss 32.04217529296875\n[180/600] Style Loss 34.1079, Content Loss 31.90805435180664\n[190/600] Style Loss 37.2641, Content Loss 31.57277488708496\n[200/600] Style Loss 27.3096, Content Loss 31.456771850585938\n[210/600] Style Loss 24.5254, Content Loss 31.218570709228516\n[220/600] Style Loss 22.1505, Content Loss 30.969505310058594\n[230/600] Style Loss 20.1192, Content Loss 30.759197235107422\n[240/600] Style Loss 18.0730, Content Loss 30.493648529052734\n[250/600] Style Loss 16.4035, Content Loss 30.251787185668945\n[260/600] Style Loss 15.0762, Content Loss 29.994197845458984\n[270/600] Style Loss 13.7020, Content Loss 29.754240036010742\n[280/600] Style Loss 12.5971, Content Loss 29.49911117553711\n[290/600] Style Loss 11.6450, Content Loss 29.278596878051758\n[300/600] Style Loss 10.8885, Content Loss 29.027385711669922\n[310/600] Style Loss 10.3600, Content Loss 28.788755416870117\n[320/600] Style Loss 9.4420, Content Loss 28.623498916625977\n[330/600] Style Loss 8.8160, Content Loss 28.381839752197266\n[340/600] Style Loss 8.2037, Content Loss 28.1502628326416\n[350/600] Style Loss 7.7651, Content Loss 27.919567108154297\n[360/600] Style Loss 7.2097, Content Loss 27.767620086669922\n[370/600] Style Loss 6.7861, Content Loss 27.577590942382812\n[380/600] Style Loss 6.4192, Content Loss 27.3933162689209\n[390/600] Style Loss 6.0287, Content Loss 27.208782196044922\n[400/600] Style Loss 5.7178, Content Loss 27.02840805053711\n[410/600] Style Loss 5.4190, Content Loss 26.81221580505371\n[420/600] Style Loss 5.0295, Content Loss 26.66666030883789\n[430/600] Style Loss 4.7181, Content Loss 26.474632263183594\n[440/600] Style Loss 4.4720, Content Loss 26.35723876953125\n[450/600] Style Loss 4.2306, Content Loss 26.198747634887695\n[460/600] Style Loss 4.0172, Content Loss 26.03799819946289\n[470/600] Style Loss 3.7802, Content Loss 25.899450302124023\n[480/600] Style Loss 4.3728, Content Loss 25.672344207763672\n[490/600] Style Loss 3.3955, Content Loss 25.647024154663086\n[500/600] Style Loss 3.2563, Content Loss 25.517745971679688\n[510/600] Style Loss 3.1030, Content Loss 25.393512725830078\n[520/600] Style Loss 3.0081, Content Loss 25.26365852355957\n[530/600] Style Loss 2.8389, Content Loss 25.16840362548828\n[540/600] Style Loss 2.7011, Content Loss 25.061481475830078\n[550/600] Style Loss 2.5967, Content Loss 24.921926498413086\n[560/600] Style Loss 2.4828, Content Loss 24.828998565673828\n[570/600] Style Loss 2.3790, Content Loss 24.728757858276367\n[580/600] Style Loss 2.2858, Content Loss 24.615867614746094\n[590/600] Style Loss 2.2063, Content Loss 24.523954391479492\n[600/600] Style Loss 2.1230, Content Loss 24.429014205932617\n[610/600] Style Loss 2.0673, Content Loss 24.335756301879883\n[620/600] Style Loss 1.9885, Content Loss 24.26019287109375\n"
]
],
[
[
"## Results",
"_____no_output_____"
]
],
[
[
"x = [r.run for r in results]\ny1 = [r.style for r in results]\ny2 = [r.content for r in results]\n\nfig, ax1 = plt.subplots(figsize=(10, 5))\n\ncolor = 'tab:red'\nax1.plot(x, y1, color=color)\nax1.set_ylabel('Style Loss', color=color)\nax1.tick_params(axis='y', labelcolor=color)\n\ncolor = 'tab:blue'\nax2 = ax1.twinx()\nax2.plot(x, y2, color=color)\nax2.set_ylabel('Content Loss', color=color)\nax2.tick_params(axis='y', labelcolor=color)",
"_____no_output_____"
]
],
[
[
"## Visualize",
"_____no_output_____"
]
],
[
[
"plt.figure()\nimshow(output, title='Output Image')\n\n# sphinx_gallery_thumbnail_number = 4\nplt.ioff()\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b6d9d1bf35470049b025d634f7435ef9e4007f | 11,833 | ipynb | Jupyter Notebook | lab work/Lab5--Practice Querying real world datasets.ipynb | Mstoned/Database-and-SQL | 066d1f65e2cfdcdbc4215a05d8b09dc7fc426a66 | [
"MIT"
] | 2 | 2020-04-03T03:27:45.000Z | 2021-04-06T13:02:21.000Z | lab work/Lab5--Practice Querying real world datasets.ipynb | Mstoned/Database-and-SQL | 066d1f65e2cfdcdbc4215a05d8b09dc7fc426a66 | [
"MIT"
] | null | null | null | lab work/Lab5--Practice Querying real world datasets.ipynb | Mstoned/Database-and-SQL | 066d1f65e2cfdcdbc4215a05d8b09dc7fc426a66 | [
"MIT"
] | null | null | null | 31.554667 | 353 | 0.62275 | [
[
[
"# Lab: Working with a real world data-set using SQL and Python",
"_____no_output_____"
],
[
"## Introduction\n\nThis notebook shows how to work with a real world dataset using SQL and Python. In this lab you will:\n1. Understand the dataset for Chicago Public School level performance \n1. Store the dataset in an Db2 database on IBM Cloud instance\n1. Retrieve metadata about tables and columns and query data from mixed case columns\n1. Solve example problems to practice your SQL skills including using built-in database functions\n\n## Chicago Public Schools - Progress Report Cards (2011-2012) \n\nThe city of Chicago released a dataset showing all school level performance data used to create School Report Cards for the 2011-2012 school year. The dataset is available from the Chicago Data Portal: https://data.cityofchicago.org/Education/Chicago-Public-Schools-Progress-Report-Cards-2011-/9xs2-f89t\n\nThis dataset includes a large number of metrics. Start by familiarizing yourself with the types of metrics in the database: https://data.cityofchicago.org/api/assets/AAD41A13-BE8A-4E67-B1F5-86E711E09D5F?download=true\n\n__NOTE__: Do not download the dataset directly from City of Chicago portal. Instead download a more database friendly version from the link below.\nNow download a static copy of this database and review some of its contents:\nhttps://ibm.box.com/shared/static/0g7kbanvn5l2gt2qu38ukooatnjqyuys.csv\n",
"_____no_output_____"
],
[
"### Store the dataset in a Table\nIn many cases the dataset to be analyzed is available as a .CSV (comma separated values) file, perhaps on the internet. To analyze the data using SQL, it first needs to be stored in the database.\n\nWhile it is easier to read the dataset into a Pandas dataframe and then PERSIST it into the database as we saw in the previous lab, it results in mapping to default datatypes which may not be optimal for SQL querying. For example a long textual field may map to a CLOB instead of a VARCHAR. \n\nTherefore, __it is highly recommended to manually load the table using the database console LOAD tool, as indicated in Week 2 Lab 1 Part II__. The only difference with that lab is that in Step 5 of the instructions you will need to click on create \"(+) New Table\" and specify the name of the table you want to create and then click \"Next\". \n\n##### Now open the Db2 console, open the LOAD tool, Select / Drag the .CSV file for the CHICAGO PUBLIC SCHOOLS dataset and load the dataset into a new table called __SCHOOLS__.\n\n<a href=\"https://cognitiveclass.ai\"><img src = \"https://ibm.box.com/shared/static/uc4xjh1uxcc78ks1i18v668simioz4es.jpg\"></a>",
"_____no_output_____"
],
[
"### Connect to the database\nLet us now load the ipython-sql extension and establish a connection with the database",
"_____no_output_____"
]
],
[
[
"%load_ext sql",
"_____no_output_____"
],
[
"# Enter the connection string for your Db2 on Cloud database instance below\n# %sql ibm_db_sa://my-username:my-password@my-hostname:my-port/my-db-name\n%sql ibm_db_sa://",
"_____no_output_____"
]
],
[
[
"### Query the database system catalog to retrieve table metadata\n\n#### You can verify that the table creation was successful by retrieving the list of all tables in your schema and checking whether the SCHOOLS table was created",
"_____no_output_____"
]
],
[
[
"# type in your query to retrieve list of all tables in the database for your db2 schema (username)\n\n#In Db2 the system catalog table called SYSCAT.TABLES contains the table metadata\n%sql SELECT * from SYSCAT.TABLES where TABNAME = 'SCHOOLS'\n\n#OR\n%sql select TABSCHEMA, TABNAME, CREATE_TIME from SYSCAT.TABLES where TABSCHEMA='YOUR-DB2-USERNAME'\n\n#OR\n%sql select TABSCHEMA, TABNAME, CREATE_TIME from SYSCAT.TABLES \\\n where TABSCHEMA not in ('SYSIBM', 'SYSCAT', 'SYSSTAT', 'SYSIBMADM', 'SYSTOOLS', 'SYSPUBLIC')",
"_____no_output_____"
]
],
[
[
"### Query the database system catalog to retrieve column metadata\n\n#### The SCHOOLS table contains a large number of columns. How many columns does this table have?",
"_____no_output_____"
]
],
[
[
"#In Db2 the system catalog table called SYSCAT.COLUMNS contains the column metadata\n%sql SELECT COUNT(*) FROM SYSCAT.COLUMNS WHERE TABNAME = 'SCHOOLS'\n\n#Correct answer: 78",
"_____no_output_____"
]
],
[
[
"Now retrieve the the list of columns in SCHOOLS table and their column type (datatype) and length.",
"_____no_output_____"
]
],
[
[
"%sql SELECT COLNAME, TYPENAME, LENGTH FROM SYSCAT.COLUMNS WHERE TABNAME = 'SCHOOLS'\n\n#OR\n%sql SELECT DISTINCT(NAME), COLTYPE, LENGTH FROM SYSIBM.SYSCOLUMNS WHERE TABNAME = 'SCHOOLS'",
"_____no_output_____"
]
],
[
[
"### Questions\n1. Is the column name for the \"SCHOOL ID\" attribute in upper or mixed case?\n1. What is the name of \"Community Area Name\" column in your table? Does it have spaces?\n1. Are there any columns in whose names the spaces and paranthesis (round brackets) have been replaced by the underscore character \"_\"?",
"_____no_output_____"
],
[
"## Problems\n\n### Problem 1\n\n##### How many Elementary Schools are in the dataset?",
"_____no_output_____"
]
],
[
[
"%sql select count(*) from SCHOOLS where \"Elementary, Middle, or High School\" = 'ES'\n\n#Correct answer: 462",
"_____no_output_____"
]
],
[
[
"### Problem 2\n\n##### What is the highest Safety Score?",
"_____no_output_____"
]
],
[
[
"%sql SELECT MAX(\"Safety_Score\") AS MAX_SAFETY FROM SCHOOLS\n\n#Correct answer: 99",
"_____no_output_____"
]
],
[
[
"### Problem 3\n\n##### Which schools have highest Safety Score?",
"_____no_output_____"
]
],
[
[
"%sql SELECT NAME_OF_SCHOOL FROM SCHOOLS WHERE \"Safety_Score\" = 99\n\n#OR\n%sql SELECT NAME_OF_SCHOOL FROM SCHOOLS WHERE \"Safety_Score\" = (SELECT MAX(\"Safety_Score\") FROM SCHOOLS)",
"_____no_output_____"
]
],
[
[
"### Problem 4\n\n##### What are the top 10 schools with the highest \"Average Student Attendance\"?",
"_____no_output_____"
]
],
[
[
"%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS ORDER BY Average_Student_Attendance DESC LIMIT 10",
"_____no_output_____"
]
],
[
[
"### Problem 5\n\n#### Retrieve the list of 5 Schools with the lowest Average Student Attendance sorted in ascending order based on attendance",
"_____no_output_____"
]
],
[
[
"%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS ORDER BY Average_Student_Attendance LIMIT 5",
"_____no_output_____"
]
],
[
[
"### Problem 6\n\n#### Now remove the '%' sign from the above result set for Average Student Attendance column",
"_____no_output_____"
]
],
[
[
"%sql SELECT NAME_OF_SCHOOL, REPLACE(Average_Student_Attendance, '%', '') FROM SCHOOLS ORDER BY Average_Student_Attendance LIMIT 5",
"_____no_output_____"
]
],
[
[
"### Problem 7\n\n#### Which Schools have Average Student Attendance lower than 70%?",
"_____no_output_____"
]
],
[
[
"%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS WHERE CAST(REPLACE(Average_Student_Attendance, '%', '') AS DOUBLE) < 70\n\n#OR\n%sql SELECT NAME_OF_SCHOOL, Average_Student_Attendance FROM SCHOOLS WHERE DECIMAL(REPLACE(Average_Student_Attendance, '%', '')) < 70 ORDER BY Average_Student_Attendance",
"_____no_output_____"
]
],
[
[
"### Problem 8\n\n#### Get the total College Enrollment for each Community Area",
"_____no_output_____"
]
],
[
[
"%sql SELECT COMMUNITY_AREA_NAME, SUM(COLLEGE_ENROLLMENT) AS TOTAL_ENROLLMENT FROM SCHOOLS GROUP BY COMMUNITY_AREA_NAME",
"_____no_output_____"
]
],
[
[
"### Problem 9\n\n##### Get the 5 Community Areas with the least total College Enrollment sorted in ascending order ",
"_____no_output_____"
]
],
[
[
"%sql SELECT COMMUNITY_AREA_NAME, SUM(COLLEGE_ENROLLMENT) AS TOTAL_ENROLLMENT FROM SCHOOLS GROUP BY COMMUNITY_AREA_NAME ORDER BY TOTAL_ENROLLMENT LIMIT 5",
"_____no_output_____"
]
],
[
[
"## Summary\n\n#### In this lab you learned how to work with a real word dataset using SQL and Python. You learned how to query columns with spaces or special characters in their names and with mixed case names. You also used built in database functions and practiced how to sort, limit, and order result sets.",
"_____no_output_____"
],
[
"Copyright © 2018 [cognitiveclass.ai](cognitiveclass.ai?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0b6dab928cdc87b83551c94038f481ff1ba81af | 14,822 | ipynb | Jupyter Notebook | python/.ipynb_checkpoints/0107_search_sort-checkpoint.ipynb | gilwoong-kang/education.cloudsecurity | b64c782017a06db9c312e04011b53a8cf058fa49 | [
"Apache-2.0"
] | null | null | null | python/.ipynb_checkpoints/0107_search_sort-checkpoint.ipynb | gilwoong-kang/education.cloudsecurity | b64c782017a06db9c312e04011b53a8cf058fa49 | [
"Apache-2.0"
] | null | null | null | python/.ipynb_checkpoints/0107_search_sort-checkpoint.ipynb | gilwoong-kang/education.cloudsecurity | b64c782017a06db9c312e04011b53a8cf058fa49 | [
"Apache-2.0"
] | 1 | 2021-06-29T09:46:23.000Z | 2021-06-29T09:46:23.000Z | 24.869128 | 158 | 0.425246 | [
[
[
"# 검색\n\nwihle loop 를 이용한 선형 검색",
"_____no_output_____"
]
],
[
[
"from typing import Any,List\n\ndef linear_search_while(lst:List, value:Any) -> int:\n i = 0\n while i != len(lst) and lst[i] != value:\n i += 1\n if i == len(lst):\n return -1\n else:\n return 1",
"_____no_output_____"
],
[
"l = [1,2,3,4,5,6,7,8,9]\nlinear_search_while(l,9)",
"_____no_output_____"
],
[
"def linear_search_for(lst:List, value:Any) -> int:\n for i in lst:\n if lst[i] == value:\n return 1\n return -1",
"_____no_output_____"
],
[
"l = [1,2,3,4,5,6,7,8,9]\nlinear_search_for(l,9)",
"_____no_output_____"
],
[
"def linear_search_sentinal(lst:List, value:Any) -> int:\n lst.append(value)\n \n i=0\n \n while lst[i] != value:\n i += 1\n \n lst.pop()\n \n if i == len(lst):\n return -1\n else:\n return 1",
"_____no_output_____"
],
[
"l = [1,2,3,4,5,6,7,8,9]\nlinear_search_sentinal(l,9)",
"_____no_output_____"
],
[
"import time\nfrom typing import Callable, Any\n\ndef time_it(search: Callable[[list,Any],Any],L:list,v:Any):\n t1 = time.perf_counter()\n search(L,v)\n t2 = time.perf_counter()\n return (t2-t1) *1000.0",
"_____no_output_____"
],
[
"l = [1,2,3,4,5,6,7,8,9]\ntime_it(linear_search_while,l,5)",
"_____no_output_____"
]
],
[
[
"## 이진 검색\n\n반절씩 줄여나가며 탐색하는 방법",
"_____no_output_____"
]
],
[
[
"def binary_search(lst:list,value:Any) -> int:\n i = 0\n j = len(lst)-1\n \n while i != j+1:\n m = (i+j)//2\n if lst[m]<v:\n i = m+1\n else:\n j=m-1\n if 0<= i< len(lst) and lst[i]==i:\n return i\n else :\n return -1\n ",
"_____no_output_____"
],
[
"if __name__ == '__main__':\n import doctest\n doctest.testmod()",
"_____no_output_____"
]
],
[
[
"## Selection sort - 선택정렬\n\n정렬되지 않은 부분 전체를 순회하며 가장 작은 값을 찾아 정렬된 부분 우측에 위치시킨다. 이것을 모든 값이 정렬될 때까지 반복한다. n길이의 선형 자료형을 n번 반복하게 되므로 n^2",
"_____no_output_____"
]
],
[
[
"def selection_sort(l:list):\n for i in range(len(l)):\n idx = l.index(min(l[i:]),i)\n dummy = l[i]\n l[i] = l[idx]\n l[idx] = dummy\n return l",
"_____no_output_____"
],
[
"l = [7,16,3,25,2,6,1,7,3]\nprint(selection_sort(l))",
"[1, 2, 3, 3, 6, 7, 7, 16, 25]\n"
]
],
[
[
"## Insertion sort - 삽입정렬\n전체를 순회하며 현재 값이 정렬된 부분에서 올바른 위치에 삽입하는 방식.",
"_____no_output_____"
]
],
[
[
"# 기 정렬된 영역에 L[:b+1] 내 올바른 위치에 L[b]를 삽입\ndef insert(L: list, b: int) -> None:\n i = b\n while i != 0 and L[i - 1] >= L[b]:\n i = i - 1\n\n value = L[b]\n del L[b]\n L.insert(i, value)\n\ndef insertion_sort(L: list) -> None:\n i = 0\n\n while i != len(L):\n insert(L, i) \n i = i + 1\n\nL = [ 3, 4, 6, -1, 2, 5 ]\nprint(L)\ninsertion_sort(L)\nprint(L) \n",
"[3, 4, 6, -1, 2, 5]\n[-1, 2, 3, 4, 5, 6]\n"
]
],
[
[
"## Merge sort - 병합정렬",
"_____no_output_____"
]
],
[
[
"# 2개의 리스트를 하나의 정렬된 리스트로 반환\ndef merge(L1: list, L2: list) -> list:\n\n newL = [] \n i1 = 0\n i2 = 0\n\n # [ 1, 1, 2, 3, 4, 5, 6, 7 ]\n # [ 1, 3, 4, 6 ] [ 1, 2, 5, 7 ]\n # i1 \n # i2 \n while i1 != len(L1) and i2 != len(L2):\n if L1[i1] <= L2[i2]:\n newL.append(L1[i1])\n i1 += 1\n else:\n newL.append(L2[i2])\n i2 += 1\n\n newL.extend(L1[i1:])\n newL.extend(L2[i2:])\n\n return newL\n\n\ndef merge_sort(L: list) -> None: # [ 1, 3, 4, 6, 1, 2, 5, 7 ]\n workspace = []\n for i in range(len(L)): \n workspace.append([L[i]]) # [ [1], [3], [4], [6], [1], [2], [5], [7] ]\n\n i = 0\n while i < len(workspace) - 1:\n L1 = workspace[i] # [ [1], [3], [4], [6], [1], [2], [5], [7], [1,3],[4,6],[1,2],[5,7], [1,3,4,6],[1,2,5,7],[1,1,2,3,4,5,6,7] ]\n L2 = workspace[i + 1]\n newL = merge(L1, L2) \n workspace.append(newL)\n i += 2\n\n if len(workspace) != 0:\n L[:] = workspace[-1][:]\n\n\nimport time, random\n\ndef built_in(L: list) -> None:\n L.sort()\n\ndef print_times(L: list) -> None:\n print(len(L), end='\\t')\n for func in (selection_sort, insertion_sort, merge_sort, built_in):\n if func in (selection_sort, insertion_sort, merge_sort) and len(L) > 10000:\n continue\n\n L_copy = L[:]\n t1 = time.perf_counter()\n func(L_copy)\n t2 = time.perf_counter()\n print(\"{0:7.1f}\".format((t2 - t1) * 1000.0), end=\"\\t\")\n\n print()\n\nfor list_size in [ 10, 1000, 2000, 3000, 4000, 5000, 10000 ]: \n L = list(range(list_size))\n random.shuffle(L)\n print_times(L)\n",
"10\t 0.0\t 0.0\t 0.0\t 0.0\t\n1000\t 16.5\t 37.0\t 4.1\t 0.1\t\n2000\t 54.9\t 141.1\t 12.2\t 0.2\t\n3000\t 130.2\t 321.6\t 15.0\t 0.4\t\n4000\t 217.9\t 592.7\t 20.7\t 0.5\t\n5000\t 357.6\t 871.0\t 26.0\t 0.7\t\n10000\t 1450.2\t 3544.8\t 55.7\t 1.5\t\n"
]
],
[
[
"# 객체지향 프로그래밍\n\n```isinstance(object,class)``` 해당 객체가 클래스에 해당하는지 아닌지를 반환.",
"_____no_output_____"
]
],
[
[
"from typing import List,Any\n\nclass Book:\n \n \n def num_authors(self) -> int:\n return len(self.authors)\n \n \n def __init__(self,title:str,authors:List[str],publisher:str,isbn:str,price:float) : # 생성자. \n self.title = title\n self.authors = authors[:] # [:] 를 적지 않고 직접 넘겨주면 참조형식이기 때문에 외부에서 값이 바뀌면 해당 값도 바뀜. 때문에 새로 만들어서 복사하는 방법을 채택. \n self.publisher = publisher\n self.isbn = isbn\n self.price = price\n\n def print_authors(self) -> None:\n for authors in self.authors:\n print(authors)\n def __str__(self) -> str:\n return 'Title : {}\\nAuthors : {}'.format(self.title,self.authors)\n\n def __eq__(self,other:Any) -> bool:\n if isinstance(other,Book):\n return True if self.isbn == other.isbn else False\n return False",
"_____no_output_____"
],
[
"book = Book('My book',['aaa','bbb','ccc'],'한빛출판사','123-456-789','300000.0')\nbook.print_authors()\nprint(book.num_authors())\nprint(book)\n\nnewBook = Book('My book',['aaa','bbb','ccc'],'한빛출판사','123-456-789','300000.0')\nprint(book==newBook)\n\n",
"aaa\nbbb\nccc\n3\nTitle : My book\nAuthors : ['aaa', 'bbb', 'ccc']\nTrue\n"
]
],
[
[
"레퍼런스 타입을 넘겨줄때 값을 참조하는 형식이 아닌 값을 직접 받는 형식으로 취하게 해야 한다. \n\n캡슐화 : 데이터와 그 데이터를 사용하는 코드를 한곳에 넣고 정확히 어떻게 동작하는ㄴ지 상세한 내용은 숨기는 것 \n다형성 : 하나 이상의 형태를 갖는 것. 어떤 변수를 포함하는 표현식이 변수가 참조하는 객체의 타입에 따라 서로 다른 일을 하는 것 \n상속 : 새로운 클래스는 부모 클래스(object 클래스 또는 사용자 정의 속성을 상속) ",
"_____no_output_____"
]
],
[
[
"class Member:\n def __init__(self,name:str,address:str,email:str):\n self.name = name\n self.address = address\n self.email = email\n \nclass Faculty(Member):\n def __init__(self,name:str,address:str,email:str,faculty_num:str):\n super().__init__(name,address,email)\n self.faculty_number = faculty_num\n self.courses_teaching = []",
"_____no_output_____"
],
[
"class Atom:\n '''번호, 기호, 좌표(X, Y, Z)를 갖는 원자'''\n\n def __init__(self, num: int, sym: str, x: float, y: float, z: float) -> None:\n self.num = num\n self.sym = sym\n self.center = (x, y, z)\n\n def __str__(self) -> str:\n '''(SYMBOL, X, Y, Z) 형식의 문자열을 반환'''\n return '({}, {}, {}, {}'.format(self.sym, self.center[0], self.center[1], self.center[2])\n\n def translate(self, x: float, y: float, z: float) -> None:\n self.center = (self.center[0] + x, self.center[1] + y, self.center[2] + z)\n",
"_____no_output_____"
],
[
"\nclass Molecule:\n ''' 이름과 원자 리스트를 갖는 분자 '''\n\n def __init__(self, name: str) -> None:\n self.name = name\n self.atoms = []\n\n def add(self, a: Atom) -> None:\n self.atoms.append(a)\n\n def __str__(self) -> str:\n '''(NAME, (ATOM1, ATOM2, ...)) 형식의 문자열을 반환'''\n\n atom_list = ''\n for a in self.atoms:\n atom_list = atom_list + str(a) + ', '\n \n atom_list = atom_list[:-2] # 마지막에 추가된 ', ' 문자를 제거\n\n return '({}, ({}))'.format(self.name, atom_list)\n\n def translate(self, x: float, y: float, z: float) -> None:\n for a in self.atoms:\n a.translate(x, y, z)\n\n\nammonia = Molecule(\"AMMONIA\")\nammonia.add(Atom(1, \"N\", 0.257, -0.363, 0.0))\nammonia.add(Atom(2, \"H\", 0.257, 0.727, 0.0))\nammonia.add(Atom(3, \"H\", 0.771, -0.727, 0.890))\nammonia.add(Atom(4, \"H\", 0.771, -0.727, -0.890))\nammonia.translate(0, 0, 0.2)\n\n\n\n#assert ammonia.atoms[0].center[0] == 0.257\n#assert ammonia.atoms[0].center[1] == -0.363\n\nassert ammonia.atoms[0].center[2] == 0.2\n\nprint(ammonia)\n",
"(AMMONIA, ((N, 0.257, -0.363, 0.2, (H, 0.257, 0.727, 0.2, (H, 0.771, -0.727, 1.09, (H, 0.771, -0.727, -0.69))\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
d0b6e3a4c6b70be146344d8f6b8e1fb9f3dcf06e | 4,389 | ipynb | Jupyter Notebook | tinker/mainline_cmp2brds_boot-pass_overtime.ipynb | viswanath-puttagunta/kernelinsights | 0ee212cf3db6575d928bcd06a1240e0c070a9c52 | [
"Apache-2.0"
] | null | null | null | tinker/mainline_cmp2brds_boot-pass_overtime.ipynb | viswanath-puttagunta/kernelinsights | 0ee212cf3db6575d928bcd06a1240e0c070a9c52 | [
"Apache-2.0"
] | null | null | null | tinker/mainline_cmp2brds_boot-pass_overtime.ipynb | viswanath-puttagunta/kernelinsights | 0ee212cf3db6575d928bcd06a1240e0c070a9c52 | [
"Apache-2.0"
] | null | null | null | 27.092593 | 135 | 0.538847 | [
[
[
"'''Objective: Compare 2 boards over time: Boot time and pass rate'''\nimport pandas as pd\nimport json\nfrom pandas.io.json import json_normalize\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"import requests\nfrom urlparse import urljoin\nimport kernelci_api_key\n\nBACKEND_URL = \"http://api.kernelci.org\"\nJOB = 'mainline'\nDEFCONFIG = 'multi_v7_defconfig '\nDATE_RANGE = 200\nSTORAGE_SERVER = 'http://storage.kernelci.org'\n\ndef invoke(board):\n headers = { \"Authorization\": kernelci_api_key.getkernelcikey()}\n params = {\n \"job\": JOB,\n \"board\": board,\n \"defconfig\":DEFCONFIG\n }\n url = urljoin(BACKEND_URL, \"/boot\")\n response = requests.get(url, params=params, headers=headers)\n \n #convert string resp into json, then into DataFrame\n contentjs = json.loads(response.content)\n df = json_normalize(contentjs['result'])\n \n #only keep columns of interest and rename some columns\n df2 = df[[u'_id.$oid',u'boot_result_description',u'created_on.$date',u'git_describe',u'lab_name',u'status',u'time.$date']]\n df2.columns = [u'boot_id',u'boot_result_description', u'created_on',u'git_describe', u'lab_name', u'status',u'boot_time']\n\n #Change created_on to DateTime. Then use this as index and sort\n df2['created_on'] = pd.to_datetime(df2['created_on'],unit='ms')\n df2.index = df2.created_on\n df2 = df2.sort_index()\n \n #drop rows for df2.status=OFFLINE\n df2 = df2[(df2.status == 'PASS') | (df2.status == 'FAIL')]\n \n #make new col of status in float\n df2['status_fl'] = df2.status.apply(lambda x: 1 if x=='PASS' else 0)\n\n #make new col of period. Used this to club(groupby) week\n df2['period'] = df2.created_on.apply(lambda x: pd.Period(x, 'W'))\n df2 = df2.groupby('period').mean()\n \n #Add boardname to col names so DataFrames can be merged later\n cols = [board+'_'+ x for x in df2.columns]\n df2.columns = cols\n \n return df2",
"_____no_output_____"
],
[
"BOARD_1 = 'am335x-boneblack'\nBOARD_2 = 'hisi-x5hd2-dkb'\ndf1 = invoke(BOARD_1)\ndf2 = invoke(BOARD_2)",
"_____no_output_____"
],
[
"df3 = df1.join(df2, how='inner')\ndf3.head()",
"_____no_output_____"
],
[
"df3[BOARD_1 + '_boot_time'].plot()\ndf3[BOARD_2 + '_boot_time'].plot()\nplt.legend(['y='+BOARD_1,'y='+BOARD_2], loc='upper right')",
"_____no_output_____"
],
[
"df3[BOARD_1 + '_status_fl'].plot()\ndf3[BOARD_2 + '_status_fl'].plot()\nplt.legend(['y='+BOARD_1,'y='+BOARD_2], loc='lower right')\nplt.ylim(0.4,1)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b6e44f53125541943d2dfc61507666731c5a59 | 60,264 | ipynb | Jupyter Notebook | EmployeeSQL/EmployeePy.ipynb | luiserojas74/sql-challenge | a8b076469649e299d655b5ac4a48fdb0ac76e7e0 | [
"ADSL"
] | null | null | null | EmployeeSQL/EmployeePy.ipynb | luiserojas74/sql-challenge | a8b076469649e299d655b5ac4a48fdb0ac76e7e0 | [
"ADSL"
] | null | null | null | EmployeeSQL/EmployeePy.ipynb | luiserojas74/sql-challenge | a8b076469649e299d655b5ac4a48fdb0ac76e7e0 | [
"ADSL"
] | null | null | null | 67.109131 | 21,484 | 0.708483 | [
[
[
"## Steps to generate a visualization of the data:",
"_____no_output_____"
],
[
"- Import the SQL database into Pandas.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sqlalchemy import create_engine\nfrom config import ppwd\nimport matplotlib.pyplot as plt\nimport scipy.stats as sts",
"_____no_output_____"
]
],
[
[
"- Configure connection to postgres",
"_____no_output_____"
]
],
[
[
"username='postgres'\npassword=ppwd\nport=5432\ndatabase='Employee_DB'\nconnection_str = f\"postgresql://{username}:{password}@localhost:{port}/{database}\"\nengine = create_engine(connection_str)\nconnection = engine.connect()",
"_____no_output_____"
]
],
[
[
"- Read tables from postgres",
"_____no_output_____"
]
],
[
[
"titles_df = pd.read_sql(\"SELECT * FROM titles\", connection)\ntitles_df",
"_____no_output_____"
],
[
"salaries_df = pd.read_sql(\"SELECT * FROM salaries\", connection)\nsalaries_df",
"_____no_output_____"
],
[
"employees_df = pd.read_sql(\"SELECT * FROM employees\", connection)\nemployees_df",
"_____no_output_____"
]
],
[
[
"- Create a histogram to visualize the most common salary ranges for employees.",
"_____no_output_____"
]
],
[
[
"salaries=salaries_df['salary']\nplt.hist(salaries)\nplt.title('Most common salary ranges for employees')\nplt.xlabel('Salaries')\nplt.ylabel('Counts')\nplt.show()",
"_____no_output_____"
]
],
[
[
"- Join three tables",
"_____no_output_____"
]
],
[
[
"merged_df = pd.merge(employees_df, salaries_df, on=\"emp_no\")\nmerged_df = pd.merge(merged_df, titles_df, left_on=\"emp_title_id\", right_on=\"title_id\")\nmerged_df",
"_____no_output_____"
]
],
[
[
"- Determine average salary by title",
"_____no_output_____"
]
],
[
[
"avg_salary_df = merged_df.groupby(\"title\").mean()['salary'].round(1).reset_index()\navg_salary_df",
"_____no_output_____"
]
],
[
[
"- Create a bar chart of average salary by title.",
"_____no_output_____"
]
],
[
[
"y_axis = avg_salary_df['salary']\nx_axis = avg_salary_df['title']\nplt.xticks(rotation=60)\nplt.title('Average salary by title')\nplt.xlabel('Title')\nplt.ylabel('Avg. Salary')\nmy_plt=plt.bar(x_axis, y_axis, color='g', alpha=0.5, align=\"center\")",
"_____no_output_____"
]
],
[
[
"- ### Managers earn almost the same as other positions. Additionally, Senior staff and Staff have no difference in salary.\n- ### According to this visualization, it seems that we were handed spurious data in order to test the data engineering skills of a new employee.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b6e855f1b88a06d516671872ce7d81c14cd774 | 16,517 | ipynb | Jupyter Notebook | IBM Professional Certificates/Databases and SQL for Data Science with Python/3-1-2-Querying.ipynb | Bezhuang/LearnCS | d82cc691e1854454576c769a090f4cfb5f91c47a | [
"MIT"
] | 1 | 2021-10-03T05:14:25.000Z | 2021-10-03T05:14:25.000Z | IBM Professional Certificates/Databases and SQL for Data Science with Python/3-1-2-Querying.ipynb | Bezhuang/LearnCS | d82cc691e1854454576c769a090f4cfb5f91c47a | [
"MIT"
] | null | null | null | IBM Professional Certificates/Databases and SQL for Data Science with Python/3-1-2-Querying.ipynb | Bezhuang/LearnCS | d82cc691e1854454576c769a090f4cfb5f91c47a | [
"MIT"
] | 1 | 2021-05-25T03:04:08.000Z | 2021-05-25T03:04:08.000Z | 30.587037 | 1,399 | 0.586305 | [
[
[
"<center>\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/Logos/organization_logo/organization_logo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n\n# Access DB2 on Cloud using Python\n\nEstimated time needed: **15** minutes\n\n## Objectives\n\nAfter completing this lab you will be able to:\n\n- Create a table\n- Insert data into the table\n- Query data from the table\n- Retrieve the result set into a pandas dataframe\n- Close the database connection\n",
"_____no_output_____"
],
[
"**Notice:** Please follow the instructions given in the first Lab of this course to Create a database service instance of Db2 on Cloud.\n\n## Task 1: Import the `ibm_db` Python library\n\nThe `ibm_db` [API ](https://pypi.python.org/pypi/ibm_db?cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ&cm_mmc=Email_Newsletter-_-Developer_Ed%2BTech-_-WW_WW-_-SkillsNetwork-Courses-IBMDeveloperSkillsNetwork-DB0201EN-SkillsNetwork-20127838&cm_mmca1=000026UJ&cm_mmca2=10006555&cm_mmca3=M12345678&cvosrc=email.Newsletter.M12345678&cvo_campaign=000026UJ) provides a variety of useful Python functions for accessing and manipulating data in an IBM® data server database, including functions for connecting to a database, preparing and issuing SQL statements, fetching rows from result sets, calling stored procedures, committing and rolling back transactions, handling errors, and retrieving metadata.\n\nWe import the ibm_db library into our Python Application\n",
"_____no_output_____"
]
],
[
[
"import ibm_db",
"_____no_output_____"
]
],
[
[
"When the command above completes, the `ibm_db` library is loaded in your notebook. \n\n## Task 2: Identify the database connection credentials\n\nConnecting to dashDB or DB2 database requires the following information:\n\n- Driver Name\n- Database name \n- Host DNS name or IP address \n- Host port\n- Connection protocol\n- User ID\n- User Password\n\n**Notice:** To obtain credentials please refer to the instructions given in the first Lab of this course\n\nNow enter your database credentials below\n\nReplace the placeholder values in angular brackets <> below with your actual database credentials \n\ne.g. replace \"database\" with \"BLUDB\"\n",
"_____no_output_____"
]
],
[
[
"#Replace the placeholder values with the actuals for your Db2 Service Credentials\ndsn_driver = \"{IBM DB2 ODBC DRIVER}\"\ndsn_database = \"database\" # e.g. \"BLUDB\"\ndsn_hostname = \"hostname\" # e.g.: \"dashdb-txn-sbox-yp-dal09-04.services.dal.bluemix.net\"\ndsn_port = \"port\" # e.g. \"50000\" \ndsn_protocol = \"protocol\" # i.e. \"TCPIP\"\ndsn_uid = \"username\" # e.g. \"abc12345\"\ndsn_pwd = \"password\" # e.g. \"7dBZ3wWt9XN6$o0J\"",
"_____no_output_____"
]
],
[
[
"## Task 3: Create the database connection\n\nIbm_db API uses the IBM Data Server Driver for ODBC and CLI APIs to connect to IBM DB2 and Informix.\n\nCreate the database connection\n",
"_____no_output_____"
]
],
[
[
"#Create database connection\n#DO NOT MODIFY THIS CELL. Just RUN it with Shift + Enter\ndsn = (\n \"DRIVER={0};\"\n \"DATABASE={1};\"\n \"HOSTNAME={2};\"\n \"PORT={3};\"\n \"PROTOCOL={4};\"\n \"UID={5};\"\n \"PWD={6};\").format(dsn_driver, dsn_database, dsn_hostname, dsn_port, dsn_protocol, dsn_uid, dsn_pwd)\n\ntry:\n conn = ibm_db.connect(dsn, \"\", \"\")\n print (\"Connected to database: \", dsn_database, \"as user: \", dsn_uid, \"on host: \", dsn_hostname)\n\nexcept:\n print (\"Unable to connect: \", ibm_db.conn_errormsg() )\n",
"_____no_output_____"
]
],
[
[
"## Task 4: Create a table in the database\n\nIn this step we will create a table in the database with following details:\n\n<img src = \"https://ibm.box.com/shared/static/ztd2cn4xkdoj5erlk4hhng39kbp63s1h.jpg\" align=\"center\">\n",
"_____no_output_____"
]
],
[
[
"#Lets first drop the table INSTRUCTOR in case it exists from a previous attempt\ndropQuery = \"drop table INSTRUCTOR\"\n\n#Now execute the drop statment\ndropStmt = ibm_db.exec_immediate(conn, dropQuery)",
"_____no_output_____"
]
],
[
[
"## Dont worry if you get this error:\n\nIf you see an exception/error similar to the following, indicating that INSTRUCTOR is an undefined name, that's okay. It just implies that the INSTRUCTOR table does not exist in the table - which would be the case if you had not created it previously.\n\nException: [IBM][CLI Driver][DB2/LINUXX8664] SQL0204N \"ABC12345.INSTRUCTOR\" is an undefined name. SQLSTATE=42704 SQLCODE=-204\n",
"_____no_output_____"
]
],
[
[
"#Construct the Create Table DDL statement\ncreateQuery = \"create table INSTRUCTOR(ID INTEGER PRIMARY KEY NOT NULL, FNAME VARCHAR(20), LNAME VARCHAR(20), CITY VARCHAR(20), CCODE CHAR(2))\"\n\n#Execute the statement\ncreateStmt = ibm_db.exec_immediate(conn,createQuery)",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\ncreateQuery = \"create table INSTRUCTOR(ID INTEGER PRIMARY KEY NOT NULL, FNAME VARCHAR(20), LNAME VARCHAR(20), CITY VARCHAR(20), CCODE CHAR(2))\"\n\ncreateStmt = ibm_db.exec_immediate(conn,createQuery)\n```\n\n</details>\n",
"_____no_output_____"
],
[
"## Task 5: Insert data into the table\n\nIn this step we will insert some rows of data into the table. \n\nThe INSTRUCTOR table we created in the previous step contains 3 rows of data:\n\n<img src=\"https://ibm.box.com/shared/static/j5yjassxefrjknivfpekj7698dqe4d8i.jpg\" align=\"center\">\n\nWe will start by inserting just the first row of data, i.e. for instructor Rav Ahuja \n",
"_____no_output_____"
]
],
[
[
"#Construct the query\ninsertQuery = \"insert into INSTRUCTOR values (1, 'Rav', 'Ahuja', 'TORONTO', 'CA')\"\n\n#execute the insert statement\ninsertStmt = ibm_db.exec_immediate(conn, insertQuery)",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\ninsertQuery = \"insert into INSTRUCTOR values (1, 'Rav', 'Ahuja', 'TORONTO', 'CA')\"\n\ninsertStmt = ibm_db.exec_immediate(conn, insertQuery)\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Now use a single query to insert the remaining two rows of data\n",
"_____no_output_____"
]
],
[
[
"#Inerts the remaining two rows of data\ninsertQuery2 = \"insert into INSTRUCTOR values (2, 'Raul', 'Chong', 'Markham', 'CA'), (3, 'Hima', 'Vasudevan', 'Chicago', 'US')\"\n\n#execute the statement\ninsertStmt2 = ibm_db.exec_immediate(conn, insertQuery2)",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\ninsertQuery2 = \"insert into INSTRUCTOR values (2, 'Raul', 'Chong', 'Markham', 'CA'), (3, 'Hima', 'Vasudevan', 'Chicago', 'US')\"\n\ninsertStmt2 = ibm_db.exec_immediate(conn, insertQuery2)\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"## Task 6: Query data in the table\n\nIn this step we will retrieve data we inserted into the INSTRUCTOR table. \n",
"_____no_output_____"
]
],
[
[
"#Construct the query that retrieves all rows from the INSTRUCTOR table\nselectQuery = \"select * from INSTRUCTOR\"\n\n#Execute the statement\nselectStmt = ibm_db.exec_immediate(conn, selectQuery)\n\n#Fetch the Dictionary (for the first row only)\nibm_db.fetch_both(selectStmt)",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\n#Construct the query that retrieves all rows from the INSTRUCTOR table\nselectQuery = \"select * from INSTRUCTOR\"\n\n#Execute the statement\nselectStmt = ibm_db.exec_immediate(conn, selectQuery)\n\n#Fetch the Dictionary (for the first row only)\nibm_db.fetch_both(selectStmt)\n\n```\n\n</details>\n",
"_____no_output_____"
]
],
[
[
"#Fetch the rest of the rows and print the ID and FNAME for those rows\nwhile ibm_db.fetch_row(selectStmt) != False:\n print (\" ID:\", ibm_db.result(selectStmt, 0), \" FNAME:\", ibm_db.result(selectStmt, \"FNAME\"))",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\n#Fetch the rest of the rows and print the ID and FNAME for those rows\nwhile ibm_db.fetch_row(selectStmt) != False:\n print (\" ID:\", ibm_db.result(selectStmt, 0), \" FNAME:\", ibm_db.result(selectStmt, \"FNAME\"))\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Bonus: now write and execute an update statement that changes the Rav's CITY to MOOSETOWN \n",
"_____no_output_____"
]
],
[
[
"updateQuery = \"update INSTRUCTOR set CITY='MOOSETOWN' where FNAME='Rav'\"\nupdateStmt = ibm_db.exec_immediate(conn, updateQuery))",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nupdateQuery = \"update INSTRUCTOR set CITY='MOOSETOWN' where FNAME='Rav'\"\nupdateStmt = ibm_db.exec_immediate(conn, updateQuery))\n\n```\n\n</details>\n",
"_____no_output_____"
],
[
"## Task 7: Retrieve data into Pandas\n\nIn this step we will retrieve the contents of the INSTRUCTOR table into a Pandas dataframe\n",
"_____no_output_____"
]
],
[
[
"import pandas\nimport ibm_db_dbi",
"_____no_output_____"
],
[
"#connection for pandas\npconn = ibm_db_dbi.Connection(conn)",
"_____no_output_____"
],
[
"#query statement to retrieve all rows in INSTRUCTOR table\nselectQuery = \"select * from INSTRUCTOR\"\n\n#retrieve the query results into a pandas dataframe\npdf = pandas.read_sql(selectQuery, pconn)\n\n#print just the LNAME for first row in the pandas data frame\npdf.LNAME[0]",
"_____no_output_____"
],
[
"#print the entire data frame\npdf",
"_____no_output_____"
]
],
[
[
"Once the data is in a Pandas dataframe, you can do the typical pandas operations on it. \n\nFor example you can use the shape method to see how many rows and columns are in the dataframe\n",
"_____no_output_____"
]
],
[
[
"pdf.shape",
"_____no_output_____"
]
],
[
[
"## Task 8: Close the Connection\n\nWe free all resources by closing the connection. Remember that it is always important to close connections so that we can avoid unused connections taking up resources.\n",
"_____no_output_____"
]
],
[
[
"ibm_db.close(conn)",
"_____no_output_____"
]
],
[
[
"## Summary\n\nIn this tutorial you established a connection to a database instance of DB2 Warehouse on Cloud from a Python notebook using ibm_db API. Then created a table and insert a few rows of data into it. Then queried the data. You also retrieved the data into a pandas dataframe.\n",
"_____no_output_____"
],
[
"## Author\n\n<a href=\"https://www.linkedin.com/in/ravahuja/\" target=\"_blank\">Rav Ahuja</a>\n\n## Change Log\n\n| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | ---------- | ---------------------------------- |\n| 2020-08-28 | 2.0 | Lavanya | Moved lab to course repo in GitLab |\n\n<hr>\n\n## <h3 align=\"center\"> © IBM Corporation 2020. All rights reserved. <h3/>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
d0b70a2d4b42a4ebbbad034f3bfc5843d751bf8f | 39,462 | ipynb | Jupyter Notebook | docs/notebook_tutorial/tutorial_MT_with_forte.ipynb | bhaskar2443053/forte | 95fabd94126d45c0db07cdcc197049ed1859d228 | [
"Apache-2.0"
] | null | null | null | docs/notebook_tutorial/tutorial_MT_with_forte.ipynb | bhaskar2443053/forte | 95fabd94126d45c0db07cdcc197049ed1859d228 | [
"Apache-2.0"
] | null | null | null | docs/notebook_tutorial/tutorial_MT_with_forte.ipynb | bhaskar2443053/forte | 95fabd94126d45c0db07cdcc197049ed1859d228 | [
"Apache-2.0"
] | null | null | null | 38.726202 | 470 | 0.608332 | [
[
[
"# Building a Machine Translation System with Forte \n\n",
"_____no_output_____"
],
[
"## Overview\n\nThis tutorial will walk you through the steps to build a machine translation system with Forte. Forte allows users to breaks down complex problems into composable pipelines and enables inter-operations across tasks through a unified data format. With Forte, it's easy to compose a customized machine translation management system that is able to handle practical problems like new feature requests.\n\nIn this tutorial, you will learn:\n\n* How to read data from source\n * How to create a simple NLP pipeline\n * How to maintain and store the input data\n* How to process data in pipeline\n * How to perform sentence segmentation\n * How to annotate and query the data\n * How to translate the input text with a pre-trained model\n * How to manage multiple data objects\n* How to handle new practical requests\n * How to handle structures like HTML data\n * How to select a single data object for processing\n * How to replace the translation model with remote translation services\n * How to save and load the pipeline\n\nRun the following command to install all the required dependencies for this tutorial:",
"_____no_output_____"
]
],
[
[
"!pip install forte==0.2.0 forte.nltk transformers==4.16.2 torch==1.7.0 requests sentencepiece",
"_____no_output_____"
]
],
[
[
"## Start with the Reader \n### Overview\n\n* **How to read data from source**\n * **How to create a simple pipeline**\n * How to maintain and store the input data\n* How to process data in pipeline\n* How to handle new practical requests\n\nIn this section, you will learn\n* What is a reader and why we need it\n* How to compose a simple pipeline with a pre-built reader\n",
"_____no_output_____"
]
],
[
[
"from forte import Pipeline\nfrom forte.data.readers import TerminalReader\npipeline: Pipeline = Pipeline()",
"_____no_output_____"
]
],
[
[
"All pipelines need a reader to read and parse input data. To make our pipeline read queries from the user’s command-line terminal, use the `TerminalReader` class provided by Forte. `TerminalReader` transforms the user’s query into a DataPack object, which is a unified data format for NLP that makes it easy to connect different NLP tools together as Forte Processors.",
"_____no_output_____"
]
],
[
[
"pipeline.set_reader(TerminalReader())",
"_____no_output_____"
]
],
[
[
"To run the pipeline consisting of the single `TerminalReader`, call `process_dataset` which will return an iterator of DataPack objects. The second line in the following code snippet retrieves the first user query from the TerminalReader. ",
"_____no_output_____"
]
],
[
[
"pipeline.initialize()\ndatapack = next(pipeline.process_dataset())\nprint(datapack.text)",
"_____no_output_____"
]
],
[
[
"### DataPack\n#### Overview\n\n* **How to read data from source**\n * How to create a simple pipeline\n * **How to maintain and store the input data**\n* How to process data in pipeline\n* How to handle new practical requests\n\nIn this section, you will learn\n* What is a DataPack object and why we need it\n\nForte helps demystify data lineage and increase the traceability of how data flows along the pipeline and how features are generated to interface data to model. Similar to a cargo ship that loads and transports goods from one port to another, a data pack carries information when passing each module and updates the ontology states along the way.\n",
"_____no_output_____"
],
[
"#### DataPack and Multi-Modality\nDataPack not only supports text data but also audio and image data.\n",
"_____no_output_____"
],
[
"## Add a pre-built Forte processor to the pipeline \n### Overview\n\n* How to read data from source\n* **How to process data in pipeline**\n * **How to perform sentence segmentation**\n * How to annotate and query the data\n * How to translate the input text with a pre-trained model\n * How to manage multiple data objects\n* How to handle new practical requests\n\nIn this section, you will learn\n* What is a processor and why we need it\n* How to add a pre-built processor to the pipeline\n\nA Forte Processor takes DataPacks as inputs, processes them, and stores its outputs in DataPacks. The processors we are going to use in this section are all PackProcessors, which expect exactly one DataPack as input and store its outputs back into the same DataPack. The following two lines of code shows how a pre-built processor `NLTKSentenceSegmenter` is added to our pipeline.",
"_____no_output_____"
]
],
[
[
"from fortex.nltk.nltk_processors import NLTKSentenceSegmenter\npipeline.add(NLTKSentenceSegmenter())",
"_____no_output_____"
]
],
[
[
"When we run the pipeline, the `NLTKSentenceSegmenter` processor will split the user query into sentences and store them back to the DataPack created by TerminalReader. The code snippet below shows how to get all the sentences from the first query.\n",
"_____no_output_____"
]
],
[
[
"from ft.onto.base_ontology import Sentence",
"_____no_output_____"
],
[
"pipeline.initialize()\nfor sent in next(pipeline.process_dataset()).get(Sentence):\n print(sent.text)",
"_____no_output_____"
]
],
[
[
"### Ontology\n#### Overview\n\n* How to read data from source\n* **How to process data in pipeline**\n * How to perform sentence segmentation\n * **How to annotate and query the data**\n * How to translate the input text with a pre-trained model\n * How to manage multiple data objects\n* How to handle new practical requests\n\nIn this section, you will learn\n* What is the ontology system and why we need it\n* How to write a customized ontology and how to use it\n\n`Sentence` is a pre-defined ontology provided by Forte and it is used by `NLTKSentenceSegmenter` to annotate each sentence in text. Forte is built on top of an Ontology system, which defines the relations between NLP annotations, for example, the relation between words and documents, or between two words. This is the core for Forte. The ontology can be specified via a JSON format. And tools are provided to convert the ontology into production code (Python).\n\n\nWe can also define customized ontologies:",
"_____no_output_____"
]
],
[
[
"from dataclasses import dataclass\nfrom forte.data.ontology.top import Annotation\nfrom typing import Optional\n\n@dataclass\nclass Article(Annotation):\n\n language: Optional[str]\n\n def __init__(self, pack, begin: int, end: int):\n super().__init__(pack, begin, end)\n self.language: Optional[str] = None",
"_____no_output_____"
]
],
[
[
"Below is a simple example showing how we can query sentences through the new ontology we just create:",
"_____no_output_____"
]
],
[
[
"from forte.data import DataPack\n\nsentences = [\n \"Do you want to get better at making delicious BBQ?\",\n \"You will have the opportunity, put this on your calendar now.\",\n \"Thursday, September 22nd join World Class BBQ Champion, Tony Balay from Lonestar Smoke Rangers.\"\n]\ndatapack: DataPack = DataPack()\n\n# Add sentences to the DataPack and annotate them\nfor sentence in sentences:\n datapack.set_text(datapack.text + sentence)\n datapack.add_entry(\n Sentence(datapack, len(datapack.text) - len(sentence), len(datapack.text))\n )\n \n# Annotate the whole text with Article\narticle: Article = Article(datapack, 0, len(datapack.text))\narticle.language = \"en\"\ndatapack.add_entry(article)\n\nfor article in datapack.get(Article):\n print(f\"Article (language - {article.language}):\")\n for sentence in article.get(Sentence):\n print(sentence.text)",
"_____no_output_____"
]
],
[
[
"In our previous example, we have the following ontologies inheritance. Sentence and Article both inherit from Annotation which is used to represent text data. In Article, we have `langauge` field to represent the text language.\n",
"_____no_output_____"
],
[
"Actually, we not only supports text ontology but also audio, image and link which represent relationships between two entries.\n\n* `Annotation` is inherited by all text entries which usually has a span to retrieve partial text from the full text.\n * `Article`, as shown in our previous example, inherits annotation and contains `language` field to differentiate English and Germany. In the single DataPack example, English article has a span of English text in the DataPack. Likewise, Germany article has a span of Germany text in the DataPack. \n * `Sentence` in our example is used to break down article, and we pass sentences into MT pipeline.\n* `AudioAnnotation` is inherited by all audio entries which usually has an audio span to retrieve partial audio from the full audio.\n * `Recording` is an example subclass of `AudioAnnotation`, and it has extra `recording_class` field denoting the classes the audio belongs to.\n* `ImageAnnotation` is inherited by all image entries which usually has payload index pointing to a loaded image array.\n * `BoundingBox` is an example subclass of `ImageAnnotation`. As the picture shows, it has more inheritance relationships than other ontology classes due to the nature of CV objects. The advantage of forte ontology is that it supports complex inheritance, and users can inherit from existing ontology and add new ontology features for their needs.\n* `Link` is inherited by all link-like entries which has parent and child.\n * `RelationLink` is an example subclass of `Link`, and it has a class attribute specifying the relation type. ",
"_____no_output_____"
],
[
"## Create a Machine Translation Processor \n### Overview\n\n* How to read data from source\n* **How to process data in pipeline**\n * How to perform sentence segmentation\n * How to annotate and query the data\n * **How to translate the input text with a pre-trained model**\n * How to manage multiple data objects\n* How to handle new practical requests\n\nIn this section, you will learn\n* The basics of machine translation process\n* How to wrap a pre-trained machine translation model into a Forte processor\n\nTranslation converts a sequence of text from one language to another. In this tutorial we will use `Huggingface` Transformer model to translate input data, which consists of several steps including subword tokenization, input embedding, model inference, decoding, etc.",
"_____no_output_____"
],
[
"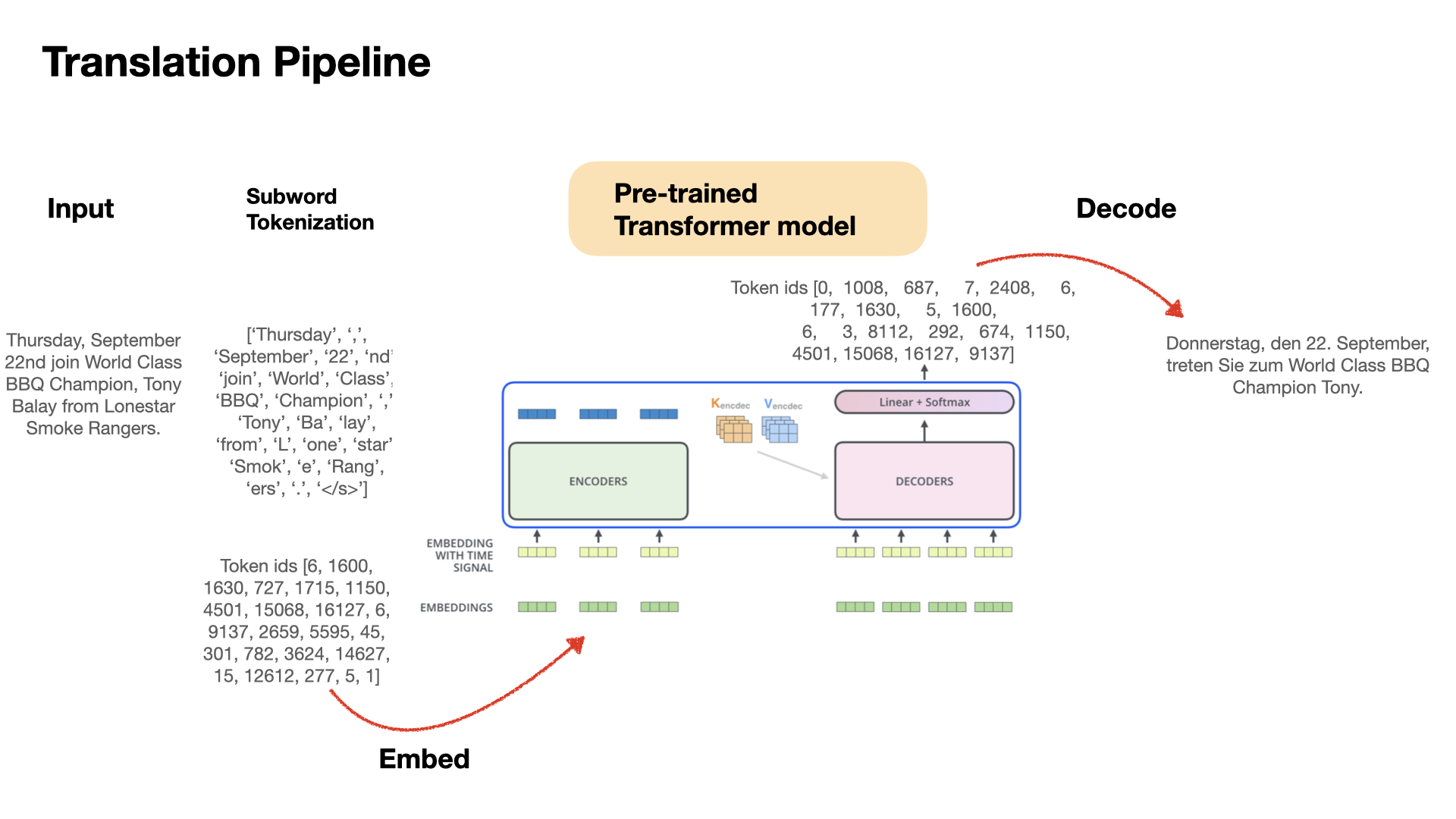",
"_____no_output_____"
],
[
"In Forte, we have a generic class `PackProcessor` that wraps model and inference-related components and behaviors to process `DataPack`. Therefore, we need to create a class that inherits the generic method from `PackProcessor`. Then we have a class definition \n`class MachineTranslationProcessor(PackProcessor)`.",
"_____no_output_____"
]
],
[
[
"from forte.data import DataPack\nfrom forte.data.readers import StringReader\nfrom forte.processors.base import PackProcessor\nfrom transformers import T5Tokenizer, T5ForConditionalGeneration\n\nclass MachineTranslationProcessor(PackProcessor):\n \"\"\"\n Translate the input text and output to a file.\n \"\"\"\n def initialize(self, resources, configs):\n super().initialize(resources, configs)\n\n # Initialize the tokenizer and model\n model_name: str = self.configs.pretrained_model\n self.tokenizer = T5Tokenizer.from_pretrained(model_name)\n self.model = T5ForConditionalGeneration.from_pretrained(model_name)\n self.task_prefix = \"translate English to German: \"\n self.tokenizer.padding_side = \"left\"\n self.tokenizer.pad_token = self.tokenizer.eos_token\n\n def _process(self, input_pack: DataPack):\n # en2de machine translation \n inputs = self.tokenizer([\n self.task_prefix + sentence.text\n for sentence in input_pack.get(Sentence)\n ], return_tensors=\"pt\", padding=True)\n\n output_sequences = self.model.generate(\n input_ids=inputs[\"input_ids\"],\n attention_mask=inputs[\"attention_mask\"],\n do_sample=False,\n )\n\n output = ''.join(self.tokenizer.batch_decode(\n output_sequences, skip_special_tokens=True\n ))\n src_article: Article = Article(input_pack, 0, len(input_pack.text))\n src_article.language = \"en\"\n\n input_pack.set_text(input_pack.text + '\\n\\n' + output)\n tgt_article: Article = Article(input_pack, len(input_pack.text) - len(output), len(input_pack.text))\n tgt_article.language = \"de\"\n\n @classmethod\n def default_configs(cls):\n return {\n \"pretrained_model\": \"t5-small\"\n }",
"_____no_output_____"
]
],
[
[
"* Initialization of needed components:\n * Users need to consider initializing all needed NLP components for the inference task such as tokenizer and model.\n * Users also need to specify all configuration in `configs`, a dictionary-like object that specifies configurations of all components such as model name. \n\n* MT operations on datapack\n * After the initialization, we already have the needed NLP components. We need to consider several MT behaviors based on Forte DataPack.\n\n * Pre-process text data\n * retrieve text data from datapack (given that it already reads data from the data source).\n * since T5 has a better performance given a task prompt, we also want to include the prompt in our data.\n\n * Tokenization that transforms input text into sequences of tokens and token ids.\n * Generate output sequences from model.\n * Decode output token ids into sentences using the tokenizer.\n\nThe generic method to process `DataPack` is `_process(self, input_pack: DataPack)`. It should tokenize the input text, use the model class to make an inference, decode the output token ids, and finally writes the output to a target file.\n\nNow we can add it into the pipeline and run the machine translation task.",
"_____no_output_____"
]
],
[
[
"input_string: str = ' '.join(sentences)\npipeline: Pipeline = Pipeline[DataPack]()\npipeline.set_reader(StringReader())\npipeline.add(NLTKSentenceSegmenter())\npipeline.add(MachineTranslationProcessor())\npipeline.initialize()\nfor datapack in pipeline.process_dataset([input_string]):\n for article in datapack.get(Article):\n print([f\"\\nArticle (language - {article.language}): {article.text}\"])",
"_____no_output_____"
]
],
[
[
"#### Ontology in DataPack\n\nHere we provide an illustration so that users can better understand the internal storage of DataPack. As we can see, text data, sentence and articles, are stored as span in `Annotations`. Their text data can be easily and efficiently retrieved by their spans.\n",
"_____no_output_____"
],
[
"## A better way to store source and target text: MultiPack \n### Overview\n\n* How to read data from source\n* **How to process data in pipeline**\n * How to perform sentence segmentation\n * How to annotate and query the data\n * How to translate the input text with a pre-trained model\n * **How to manage multiple data objects**\n* How to handle new practical requests\n\nIn this section, you will learn\n* What is a MultiPack and why we need it\n* How to use a Multipack\n\nThe above step outputs a DataPack which is good for holding data about one specific piece of text. A complicated pipeline like the one we are building now may need multiple DataPacks to be passed along the pipeline and this is where MultiPack can help. MultiPack manages a set of DataPacks that can be indexed by their names.\n\n`MultiPackBoxer` is a simple Forte processor that converts a DataPack into a MultiPack by making it the only DataPack in there. A name can be specified via the config. We use it to wrap DataPack that contains source sentence.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"from forte.data import MultiPack\nfrom forte.processors.base import MultiPackProcessor\nfrom forte.data.caster import MultiPackBoxer\n\nclass MachineTranslationMPProcessor(MultiPackProcessor):\n \"\"\"\n Translate the input text and output to a file.\n \"\"\"\n def initialize(self, resources, configs):\n super().initialize(resources, configs)\n\n # Initialize the tokenizer and model\n model_name: str = self.configs.pretrained_model\n self.tokenizer = T5Tokenizer.from_pretrained(model_name)\n self.model = T5ForConditionalGeneration.from_pretrained(model_name)\n self.task_prefix = \"translate English to German: \"\n self.tokenizer.padding_side = \"left\"\n self.tokenizer.pad_token = self.tokenizer.eos_token\n\n def _process(self, input_pack: MultiPack):\n source_pack: DataPack = input_pack.get_pack(\"source\")\n target_pack: DataPack = input_pack.add_pack(\"target\")\n\n # en2de machine translation \n inputs = self.tokenizer([\n self.task_prefix + sentence.text\n for sentence in source_pack.get(Sentence)\n ], return_tensors=\"pt\", padding=True)\n\n output_sequences = self.model.generate(\n input_ids=inputs[\"input_ids\"],\n attention_mask=inputs[\"attention_mask\"],\n do_sample=False,\n )\n \n # Annotate the source article\n src_article: Article = Article(source_pack, 0, len(source_pack.text))\n src_article.language = \"en\"\n \n # Annotate each sentence\n for output in self.tokenizer.batch_decode(\n output_sequences, skip_special_tokens=True\n ):\n target_pack.set_text(target_pack.text + output)\n text_length: int = len(target_pack.text)\n Sentence(target_pack, text_length - len(output), text_length)\n \n # Annotate the target article\n tgt_article: Article = Article(target_pack, 0, len(target_pack.text))\n tgt_article.language = \"de\"\n\n @classmethod\n def default_configs(cls):\n return {\n \"pretrained_model\": \"t5-small\",\n }",
"_____no_output_____"
]
],
[
[
"Then `MachineTranslationMPProcessor` writes the output sentence into a target DataPack.",
"_____no_output_____"
],
[
"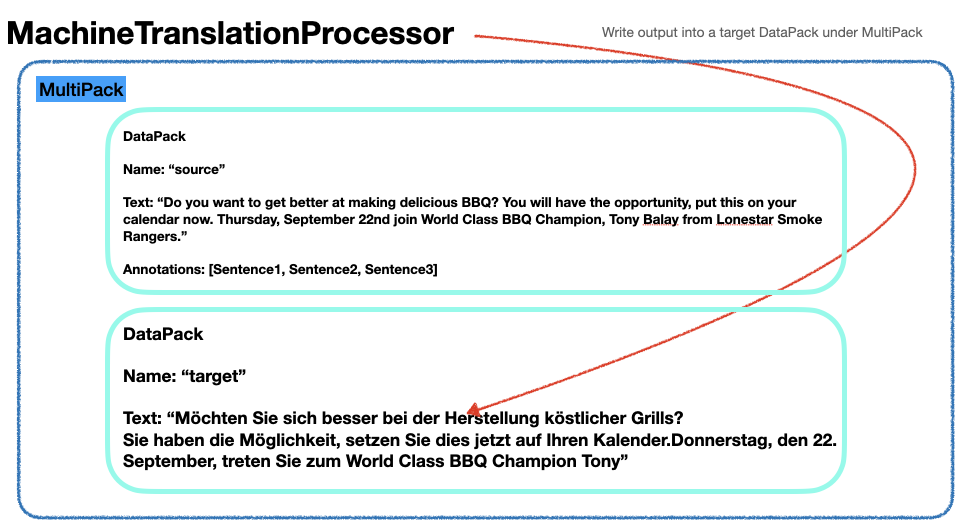",
"_____no_output_____"
],
[
"Now let's try to create a new pipeline that utilizes `MultiPack` to manage text in different languages.",
"_____no_output_____"
]
],
[
[
"nlp: Pipeline = Pipeline[DataPack]()\nnlp.set_reader(StringReader())\nnlp.add(NLTKSentenceSegmenter())\nnlp.add(MultiPackBoxer(), config={\"pack_name\": \"source\"})\nnlp.add(MachineTranslationMPProcessor(), config={\n \"pretrained_model\": \"t5-small\"\n})\nnlp.initialize()\nfor multipack in nlp.process_dataset([input_string]):\n for pack_name in (\"source\", \"target\"):\n for article in multipack.get_pack(pack_name).get(Article):\n print(f\"\\nArticle (language - {article.language}): \")\n for sentence in article.get(Sentence):\n print(sentence.text)",
"_____no_output_____"
]
],
[
[
"#### Ontology in MultiPack\n\nFor comparison, here is an illustration of the internal storage of MultiPack. We can see that MultiPack wraps one source DataPack and one target DataPack. Article spans are based on two separate DataPack text.\n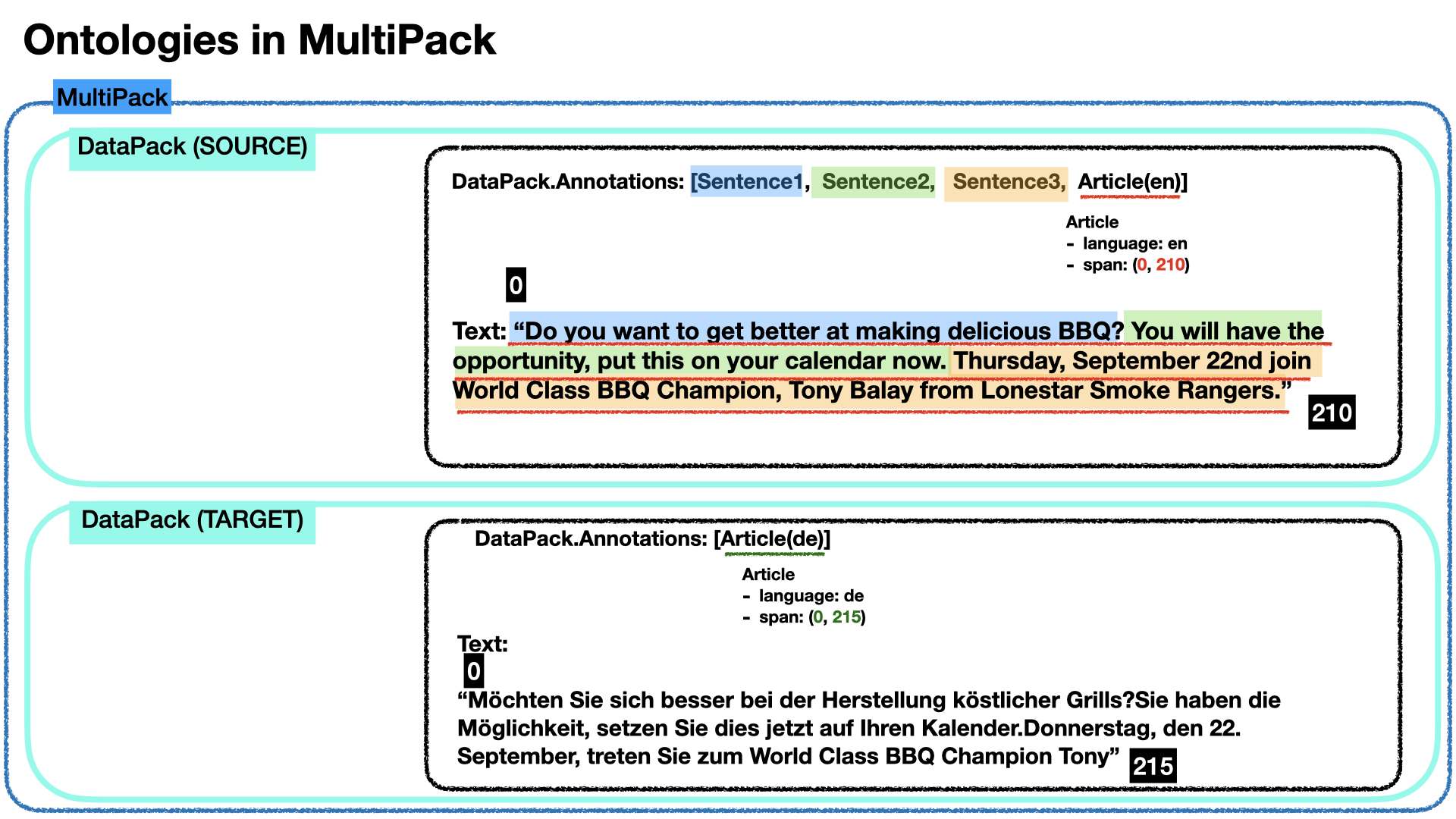",
"_____no_output_____"
],
[
"## New Requirement: Handle HTML data \n### Overview\n\n* How to read data from source\n* How to process data in pipeline\n* **How to handle new practical requests**\n * **How to handle structures like HTML data**\n * **How to select a single data object for processing**\n * How to replace the translation model with remote translation services\n * How to save and load the pipeline\n\n\nIn this section, you will learn\n* How to build a translation management system\n* How to preserve the structure like HTML in machine translation\n* How to select a specific DataPack from MultiPack for processing\n\nIn the previous step, the input string is just a simple paragraph made up of several sentences. However, in many cases, we might need to handle data with structural information, such HTML or XML. When the input is a string of raw HTML data, the machine translation pipeline above may not work as expected:",
"_____no_output_____"
]
],
[
[
"html_input: str = \"\"\"\n<!DOCTYPE html>\n<html>\n <head><title>Beginners BBQ Class.</title></head>\n <body>\n <p>Do you want to get better at making delicious BBQ? You will have the opportunity, put this on your calendar now. Thursday, September 22nd join World Class BBQ Champion, Tony Balay from Lonestar Smoke Rangers.</p>\n </body>\n</html>\n\"\"\"\nnlp.initialize()\nfor multipack in nlp.process_dataset([html_input]):\n print(\"Source Text: \" + multipack.get_pack(\"source\").text)\n print(\"\\nTarget Text: \" + multipack.get_pack(\"target\").text)",
"_____no_output_____"
]
],
[
[
"We can see that the original HTML structure is broken in the translated output.\n\n## How to preserve HTML tags/structures\n\nIn order to handle structured data like HTML, we will need to update our current design of pipeline. Luckily, Forte pipelines are highly modular, we can simply insert two new processors without updating the previous pipeline.\n\nWe first need a HTML cleaner to parse all the HTML tags from input string. Picture below shows the effect of tag remover.\n",
"_____no_output_____"
],
[
"After the translation is finished, we will also need to recover the HTML structure from the unstructured translation output. Picture below shows replace one source sentence with one target sentence given the target sentence is ready.\n",
"_____no_output_____"
]
],
[
[
"from forte.data import NameMatchSelector\nfrom forte.data.readers.html_reader import ForteHTMLParser\n\nclass HTMLTagCleaner(MultiPackProcessor):\n \n def initialize(self, resources, configs):\n super().initialize(resources, configs)\n self._parser = ForteHTMLParser()\n\n def _process(self, input_pack: MultiPack):\n raw_pack: DataPack = input_pack.get_pack(\"raw\")\n source_pack: DataPack = input_pack.add_pack(\"source\")\n \n self._parser.feed(raw_pack.text)\n cleaned_text: str = raw_pack.text\n for span, _ in self._parser.spans:\n cleaned_text = cleaned_text.replace(\n raw_pack.text[span.begin:span.end], ''\n )\n source_pack.set_text(cleaned_text)\n \nclass HTMLTagRecovery(MultiPackProcessor):\n\n def _process(self, input_pack: MultiPack):\n raw_pack: DataPack = input_pack.get_pack(\"raw\")\n source_pack: DataPack = input_pack.get_pack(\"source\")\n target_pack: DataPack = input_pack.get_pack(\"target\")\n result_pack: DataPack = input_pack.add_pack(\"result\")\n result_text: str = raw_pack.text\n for sent_src, sent_tgt in zip(source_pack.get(Sentence), target_pack.get(Sentence)):\n result_text = result_text.replace(sent_src.text, sent_tgt.text)\n result_pack.set_text(result_text)",
"_____no_output_____"
]
],
[
[
"Now we are able to create a translation management system by inserting the two processors introduced above into our previous machine translation pipeline.",
"_____no_output_____"
]
],
[
[
"# Pipeline with HTML handling\npipeline: Pipeline = Pipeline[DataPack]()\npipeline.set_reader(StringReader())\npipeline.add(MultiPackBoxer(), config={\"pack_name\": \"raw\"})\npipeline.add(HTMLTagCleaner())\npipeline.add(\n NLTKSentenceSegmenter(),\n selector=NameMatchSelector(),\n selector_config={\"select_name\": \"source\"}\n)\npipeline.add(MachineTranslationMPProcessor(), config={\n \"pretrained_model\": \"t5-small\"\n})\npipeline.add(HTMLTagRecovery())\n\npipeline.initialize()\nfor multipack in pipeline.process_dataset([html_input]):\n print(multipack.get_pack(\"raw\").text)\n print(multipack.get_pack(\"result\").text)",
"_____no_output_____"
]
],
[
[
"### Selector\nIn the code snippet above, we utilize a `NameMatchSelector` to select one specific DataPack from the MultiPack based on its reference name `select_name`. This allows `NLTKSentenceSegmenter` to process only the specified DataPack.\n\n## Replace our MT model with online translation API\n### Overview\n\n* How to read data from source\n* How to process data in pipeline\n* **How to handle new practical requests**\n * How to handle structures like HTML data\n * **How to replace the translation model with remote translation services**\n * How to save and load the pipeline\n\n\nIn this section, you will learn\n* How to use a different translation service\n\nForte also allows us to update the translation model and integrate it seamlessly to the original pipeline. For example, if we want to offload the translation task to an online service, all we need to do is to update the translation processor. There is no need to change other components in the pipeline.",
"_____no_output_____"
]
],
[
[
"# You can get your own API key by following the instructions in https://docs.microsoft.com/en-us/azure/cognitive-services/translator/\napi_key = input(\"Enter your API key here:\")",
"_____no_output_____"
],
[
"import requests\nimport uuid\n\nclass OnlineMachineTranslationMPProcessor(MultiPackProcessor):\n \"\"\"\n Translate the input text and output to a file use online translator api.\n \"\"\"\n def initialize(self, resources, configs):\n super().initialize(resources, configs)\n self.url = configs.endpoint + configs.path\n self.from_lang = configs.from_lang\n self.to_lang = configs.to_lang\n self.subscription_key = configs.subscription_key\n self.subscription_region = configs.subscription_region\n\n def _process(self, input_pack: MultiPack):\n source_pack: DataPack = input_pack.get_pack(\"source\")\n target_pack: DataPack = input_pack.add_pack(\"target\")\n \n params = {\n 'api-version': '3.0',\n 'from': 'en',\n 'to': ['de']\n }\n # Build request\n headers = {\n 'Ocp-Apim-Subscription-Key': self.subscription_key,\n 'Ocp-Apim-Subscription-Region': self.subscription_region,\n 'Content-type': 'application/json',\n 'X-ClientTraceId': str(uuid.uuid4())\n }\n # You can pass more than one object in body.\n body = [{\n 'text': source_pack.text\n }]\n\n request = requests.post(self.url, params=params, headers=headers, json=body)\n \n result = request.json()\n target_pack.set_text(\"\".join(\n [trans['text'] for trans in result[0][\"translations\"]]\n )\n )\n\n @classmethod\n def default_configs(cls):\n return {\n \"from_lang\" : 'en',\n \"to_lang\": 'de',\n \"endpoint\" : 'https://api.cognitive.microsofttranslator.com/',\n \"path\" : '/translate',\n \"subscription_key\": None,\n \"subscription_region\" : \"westus2\",\n 'X-ClientTraceId': str(uuid.uuid4())\n }",
"_____no_output_____"
],
[
"nlp: Pipeline = Pipeline[DataPack]()\nnlp.set_reader(StringReader())\nnlp.add(NLTKSentenceSegmenter())\nnlp.add(MultiPackBoxer(), config={\"pack_name\": \"source\"})\nnlp.add(OnlineMachineTranslationMPProcessor(), config={\n \"from_lang\" : 'en',\n \"to_lang\": 'de',\n \"endpoint\" : 'https://api.cognitive.microsofttranslator.com/',\n \"path\" : '/translate',\n \"subscription_key\": api_key,\n \"subscription_region\" : \"westus2\",\n 'X-ClientTraceId': str(uuid.uuid4())\n})\nnlp.initialize()\nfor multipack in nlp.process_dataset([input_string]):\n print(\"Source Text: \" + multipack.get_pack(\"source\").text)\n print(\"\\nTarget Text: \" + multipack.get_pack(\"target\").text)",
"_____no_output_____"
]
],
[
[
"## Save the whole pipeline with save() \n### Overview\n\n* How to read data from source\n* How to process data in pipeline\n* **How to handle new practical requests**\n * How to handle structures like HTML data\n * How to replace the translation model with remote translation services\n * **How to save and load the pipeline**\n\n\nIn this section, you will learn\n* How to export and import a Forte pipeline\n\nForte also allow us to save the pipeline into disk. It serializes the whole pipeline and generates an intermediate representation, which can be loaded later maybe on a different machine.",
"_____no_output_____"
]
],
[
[
"import os\nsave_path: str = os.path.join(os.path.dirname(os.path.abspath('')), \"pipeline.yml\")\nnlp.save(save_path)\n\nwith open(save_path, 'r') as f:\n print(f.read())",
"_____no_output_____"
]
],
[
[
"Now that the pipeline is saved, we can try to re-load the pipeline to see if it still functions as expect.",
"_____no_output_____"
]
],
[
[
"new_nlp: Pipeline = Pipeline()\nnew_nlp.init_from_config_path(save_path)\nnew_nlp.initialize()\nfor multipack in new_nlp.process_dataset([input_string]):\n print(\"Source Text: \" + multipack.get_pack(\"source\").text)\n print(\"\\nTarget Text: \" + multipack.get_pack(\"target\").text)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b712b785c6f07cc3ce553b844dcef3d4bf8cdc | 254,335 | ipynb | Jupyter Notebook | examples/plugin-Search.ipynb | artnikitin/folium | 4a7532d34c22532167d128dddd07b1578ed08f37 | [
"MIT"
] | 3 | 2020-01-08T18:30:07.000Z | 2021-07-25T06:54:32.000Z | examples/plugin-Search.ipynb | artnikitin/folium | 4a7532d34c22532167d128dddd07b1578ed08f37 | [
"MIT"
] | 1 | 2020-05-21T11:13:30.000Z | 2020-05-21T11:13:30.000Z | examples/plugin-Search.ipynb | artnikitin/folium | 4a7532d34c22532167d128dddd07b1578ed08f37 | [
"MIT"
] | 7 | 2019-07-21T03:30:26.000Z | 2021-12-14T04:41:27.000Z | 457.43705 | 193,692 | 0.886677 | [
[
[
"import folium\nimport branca\nimport geopandas\nfrom folium.plugins import Search\n\n\nprint(folium.__version__)",
"0.8.3+52.g2758dc7.dirty\n"
]
],
[
[
"Let's get some JSON data from the web - both a point layer and a polygon GeoJson dataset with some population data.",
"_____no_output_____"
]
],
[
[
"states = geopandas.read_file(\n 'https://rawcdn.githack.com/PublicaMundi/MappingAPI/master/data/geojson/us-states.json',\n driver='GeoJSON'\n)\n\ncities = geopandas.read_file(\n 'https://d2ad6b4ur7yvpq.cloudfront.net/naturalearth-3.3.0/ne_50m_populated_places_simple.geojson',\n driver='GeoJSON'\n)",
"_____no_output_____"
]
],
[
[
"And take a look at what our data looks like:",
"_____no_output_____"
]
],
[
[
"states.describe()",
"_____no_output_____"
]
],
[
[
"Look how far the minimum and maximum values for the density are from the top and bottom quartile breakpoints! We have some outliers in our data that are well outside the meat of most of the distribution. Let's look into this to find the culprits within the sample.",
"_____no_output_____"
]
],
[
[
"states_sorted = states.sort_values(by='density', ascending=False)\n\nstates_sorted.head(5).append(states_sorted.tail(5))[['name','density']]",
"_____no_output_____"
]
],
[
[
"Looks like Washington D.C. and Alaska were the culprits on each end of the range. Washington was more dense than the next most dense state, New Jersey, than the least dense state, Alaska was from Wyoming, however. Washington D.C. has a has a relatively small land area for the amount of people that live there, so it makes sense that it's pretty dense. And Alaska has a lot of land area, but not much of it is habitable for humans.\n<br><br>\nHowever, we're looking at all of the states in the US to look at things on a more regional level. That high figure at the top of our range for Washington D.C. will really hinder the ability for us to differentiate between the other states, so let's account for that in the min and max values for our color scale, by getting the quantile values close to the end of the range. Anything higher or lower than those values will just fall into the 'highest' and 'lowest' bins for coloring.",
"_____no_output_____"
]
],
[
[
"min, max = states['density'].quantile([0.05,0.95]).apply(lambda x: round(x, 2))\n\nmean = round(states['density'].mean(),2)\n\n\nprint(f\"Min: {min}\", f\"Max: {max}\", f\"Mean: {mean}\", sep=\"\\n\\n\")",
"Min: 8.54\n\nMax: 1040.2\n\nMean: 402.5\n"
]
],
[
[
"This looks better. Our min and max values for the colorscale are much closer to the mean value now. Let's run with these values, and make a colorscale. I'm just going to use a sequential light-to-dark color palette from the [ColorBrewer](http://colorbrewer2.org/#type=sequential&scheme=Purples&n=5).",
"_____no_output_____"
]
],
[
[
"colormap = branca.colormap.LinearColormap(\n colors=['#f2f0f7','#cbc9e2','#9e9ac8','#756bb1','#54278f'],\n index=states['density'].quantile([0.2,0.4,0.6,0.8]),\n vmin=min,\n vmax=max\n)\n\ncolormap.caption=\"Population Density in the United States\"\n\ncolormap\n",
"_____no_output_____"
]
],
[
[
"Let's narrow down these cities to United states cities, by using GeoPandas' spatial join functionality between two GeoDataFrame objects, using the Point 'within' Polygon functionality.",
"_____no_output_____"
]
],
[
[
"us_cities = geopandas.sjoin(cities, states, how='inner', op='within')\n\npop_ranked_cities = us_cities.sort_values(\n by='pop_max',\n ascending=False\n)[\n [\n 'nameascii',\n 'pop_max',\n 'geometry'\n ]\n].iloc[:20]",
"_____no_output_____"
]
],
[
[
"Ok, now we have a new GeoDataFrame with our top 20 populated cities. Let's see the top 5.",
"_____no_output_____"
]
],
[
[
"pop_ranked_cities.head(5)",
"_____no_output_____"
]
],
[
[
"Alright, let's build a map!",
"_____no_output_____"
]
],
[
[
"m = folium.Map(location=[38,-97], zoom_start=4)\n\n\nstyle_function = lambda x: {\n 'fillColor': colormap(x['properties']['density']),\n 'color': 'black',\n 'weight':2,\n 'fillOpacity':0.5\n}\n\nstategeo = folium.GeoJson(\n states,\n name='US States',\n style_function=style_function,\n tooltip=folium.GeoJsonTooltip(\n fields=['name', 'density'],\n aliases=['State', 'Density'], \n localize=True\n )\n).add_to(m)\n\ncitygeo = folium.GeoJson(\n pop_ranked_cities,\n name='US Cities',\n tooltip=folium.GeoJsonTooltip(\n fields=['nameascii','pop_max'],\n aliases=['','Population Max'],\n localize=True)\n).add_to(m)\n\nstatesearch = Search(\n layer=stategeo,\n geom_type='Polygon',\n placeholder='Search for a US State',\n collapsed=False,\n search_label='name',\n weight=3\n).add_to(m)\n\ncitysearch = Search(\n layer=citygeo,\n geom_type='Point',\n placeholder='Search for a US City',\n collapsed=True,\n search_label='nameascii'\n).add_to(m)\n\nfolium.LayerControl().add_to(m)\ncolormap.add_to(m)\n\nm",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b731fd841b4c3d1a39935dfd225397152be84a | 67,608 | ipynb | Jupyter Notebook | lectures/Week 05 - Data Processing and Visualization Part 2/02 - NumPy Data analysis.ipynb | xiangyuzeng/data-focused-python | db0b289875b1cc01744c126cf0f54d4d70d5d2af | [
"MIT"
] | 1 | 2019-09-09T14:49:55.000Z | 2019-09-09T14:49:55.000Z | lectures/Week 05 - Data Processing and Visualization Part 2/02 - NumPy Data analysis.ipynb | xiangyuzeng/data-focused-python | db0b289875b1cc01744c126cf0f54d4d70d5d2af | [
"MIT"
] | null | null | null | lectures/Week 05 - Data Processing and Visualization Part 2/02 - NumPy Data analysis.ipynb | xiangyuzeng/data-focused-python | db0b289875b1cc01744c126cf0f54d4d70d5d2af | [
"MIT"
] | null | null | null | 28.562738 | 650 | 0.527512 | [
[
[
"# NumPy Tutorial: Data analysis with Python\n[Source](https://www.dataquest.io/blog/numpy-tutorial-python/)\n\nNumPy is a commonly used Python data analysis package. By using NumPy, you can speed up your workflow, and interface with other packages in the Python ecosystem, like scikit-learn, that use NumPy under the hood. NumPy was originally developed in the mid 2000s, and arose from an even older package called Numeric. This longevity means that almost every data analysis or machine learning package for Python leverages NumPy in some way.\n\nIn this tutorial, we'll walk through using NumPy to analyze data on wine quality. The data contains information on various attributes of wines, such as pH and fixed acidity, along with a quality score between 0 and 10 for each wine. The quality score is the average of at least 3 human taste testers. As we learn how to work with NumPy, we'll try to figure out more about the perceived quality of wine.\n\nThe wines we'll be analyzing are from the Minho region of Portugal.",
"_____no_output_____"
],
[
"The data was downloaded from the UCI Machine Learning Repository, and is available [here](https://archive.ics.uci.edu/ml/datasets/Wine+Quality). Here are the first few rows of the winequality-red.csv file, which we'll be using throughout this tutorial:\n\n``` text\n\"fixed acidity\";\"volatile acidity\";\"citric acid\";\"residual sugar\";\"chlorides\";\"free sulfur dioxide\";\"total sulfur dioxide\";\"density\";\"pH\";\"sulphates\";\"alcohol\";\"quality\"\n7.4;0.7;0;1.9;0.076;11;34;0.9978;3.51;0.56;9.4;5\n7.8;0.88;0;2.6;0.098;25;67;0.9968;3.2;0.68;9.8;5\n```\n\nThe data is in what I'm going to call ssv (semicolon separated values) format -- each record is separated by a semicolon (;), and rows are separated by a new line. There are 1600 rows in the file, including a header row, and 12 columns.\n\nBefore we get started, a quick version note -- we'll be using Python 3.5. Our code examples will be done using Jupyter notebook.\n\nIf you want to jump right into a specific area, here are the topics:\n* Creating an Array\n* Reading Text Files\n* Array Indexing\n* N-Dimensional Arrays\n* Data Types\n* Array Math\n* Array Methods\n* Array Comparison and Filtering\n* Reshaping and Combining Arrays",
"_____no_output_____"
],
[
"Lists Of Lists for CSV Data\nBefore using NumPy, we'll first try to work with the data using Python and the csv package. We can read in the file using the csv.reader object, which will allow us to read in and split up all the content from the ssv file.\n\nIn the below code, we:\n\n* Import the csv library.\n* Open the winequality-red.csv file.\n * With the file open, create a new csv.reader object.\n * Pass in the keyword argument delimiter=\";\" to make sure that the records are split up on the semicolon character instead of the default comma character.\n * Call the list type to get all the rows from the file.\n * Assign the result to wines.",
"_____no_output_____"
]
],
[
[
"import csv\n\nwith open(\"winequality-red.csv\", 'r') as f:\n wines = list(csv.reader(f, delimiter=\";\"))\n# print(wines[:3])\n \nheaders = wines[0]\nwines_only = wines[1:]",
"_____no_output_____"
],
[
"# print the headers\nprint(headers)",
"['fixed acidity', 'volatile acidity', 'citric acid', 'residual sugar', 'chlorides', 'free sulfur dioxide', 'total sulfur dioxide', 'density', 'pH', 'sulphates', 'alcohol', 'quality']\n"
],
[
"# print the 1st row of data\nprint(wines_only[0])",
"['7.4', '0.7', '0', '1.9', '0.076', '11', '34', '0.9978', '3.51', '0.56', '9.4', '5']\n"
],
[
"# print the 1st three rows of data\nprint(wines_only[:3])",
"[['7.4', '0.7', '0', '1.9', '0.076', '11', '34', '0.9978', '3.51', '0.56', '9.4', '5'], ['7.8', '0.88', '0', '2.6', '0.098', '25', '67', '0.9968', '3.2', '0.68', '9.8', '5'], ['7.8', '0.76', '0.04', '2.3', '0.092', '15', '54', '0.997', '3.26', '0.65', '9.8', '5']]\n"
]
],
[
[
"The data has been read into a list of lists. Each inner list is a row from the ssv file. As you may have noticed, each item in the entire list of lists is represented as a string, which will make it harder to do computations.\n\nAs you can see from the table above, we've read in three rows, the first of which contains column headers. Each row after the header row represents a wine. The first element of each row is the fixed acidity, the second is the volatile acidity, and so on. \n\n## Calculate Average Wine Quality\n\nWe can find the average quality of the wines. The below code will:\n\n* Extract the last element from each row after the header row.\n* Convert each extracted element to a float.\n* Assign all the extracted elements to the list qualities.\n* Divide the sum of all the elements in qualities by the total number of elements in qualities to the get the mean.",
"_____no_output_____"
]
],
[
[
"# calculate average wine quality with a loop\nqualities = []\nfor row in wines[1:]:\n qualities.append(float(row[-1]))\n\nsum(qualities) / len(wines[1:])",
"_____no_output_____"
],
[
"# calculate average wine quality with a list comprehension\nqualities = [float(row[-1]) for row in wines[1:]]\n\nsum(qualities) / len(wines[1:])",
"_____no_output_____"
]
],
[
[
"Although we were able to do the calculation we wanted, the code is fairly complex, and it won't be fun to have to do something similar every time we want to compute a quantity. Luckily, we can use NumPy to make it easier to work with our data.",
"_____no_output_____"
],
[
"# Numpy 2-Dimensional Arrays\n\nWith NumPy, we work with multidimensional arrays. We'll dive into all of the possible types of multidimensional arrays later on, but for now, we'll focus on 2-dimensional arrays. A 2-dimensional array is also known as a matrix, and is something you should be familiar with. In fact, it's just a different way of thinking about a list of lists. A matrix has rows and columns. By specifying a row number and a column number, we're able to extract an element from a matrix.\n\nIf we picked the element at the first row and the second column, we'd get volatile acidity. If we picked the element in the third row and the second column, we'd get 0.88.\n\nIn a NumPy array, the number of dimensions is called the **rank**, and each dimension is called an **axis**. So \n* the rows are the first axis\n* the columns are the second axis\n\nNow that you understand the basics of matrices, let's see how we can get from our list of lists to a NumPy array.",
"_____no_output_____"
],
[
"## Creating A NumPy Array\n\nWe can create a NumPy array using the numpy.array function. If we pass in a list of lists, it will automatically create a NumPy array with the same number of rows and columns. Because we want all of the elements in the array to be float elements for easy computation, we'll leave off the header row, which contains strings. One of the limitations of NumPy is that all the elements in an array have to be of the same type, so if we include the header row, all the elements in the array will be read in as strings. Because we want to be able to do computations like find the average quality of the wines, we need the elements to all be floats.\n\nIn the below code, we:\n\n* Import the ```numpy``` package.\n* Pass the ```list``` of lists wines into the array function, which converts it into a NumPy array.\n * Exclude the header row with list slicing.\n * Specify the keyword argument ```dtype``` to make sure each element is converted to a ```float```. We'll dive more into what the ```dtype``` is later on.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnp.set_printoptions(precision=2) # set the output print precision for readability\n\n# create the numpy array skipping the headers\nwines = np.array(wines[1:], dtype=np.float)",
"_____no_output_____"
],
[
"# If we display wines, we'll now get a NumPy array:\nprint(type(wines), wines)",
"<class 'numpy.ndarray'> [[ 7.4 0.7 0. ... 0.56 9.4 5. ]\n [ 7.8 0.88 0. ... 0.68 9.8 5. ]\n [ 7.8 0.76 0.04 ... 0.65 9.8 5. ]\n ...\n [ 6.3 0.51 0.13 ... 0.75 11. 6. ]\n [ 5.9 0.65 0.12 ... 0.71 10.2 5. ]\n [ 6. 0.31 0.47 ... 0.66 11. 6. ]]\n"
],
[
"# We can check the number of rows and columns in our data using the shape property of NumPy arrays:\nwines.shape",
"_____no_output_____"
]
],
[
[
"## Alternative NumPy Array Creation Methods\n\nThere are a variety of methods that you can use to create NumPy arrays. It's useful to create an array with all zero elements in cases when you need an array of fixed size, but don't have any values for it yet. To start with, you can create an array where every element is zero. The below code will create an array with 3 rows and 4 columns, where every element is 0, using ```numpy.zeros```:",
"_____no_output_____"
]
],
[
[
"empty_array = np.zeros((3, 4))\nempty_array",
"_____no_output_____"
]
],
[
[
"Creating arrays full of random numbers can be useful when you want to quickly test your code with sample arrays. You can also create an array where each element is a random number using ```numpy.random.rand```.",
"_____no_output_____"
]
],
[
[
"np.random.rand(2, 3)",
"_____no_output_____"
]
],
[
[
"### Using NumPy To Read In Files\nIt's possible to use NumPy to directly read ```csv``` or other files into arrays. We can do this using the ```numpy.genfromtxt``` function. We can use it to read in our initial data on red wines.\n\nIn the below code, we:\n\n* Use the ``` genfromtxt ``` function to read in the ``` winequality-red.csv ``` file.\n* Specify the keyword argument ``` delimiter=\";\" ``` so that the fields are parsed properly.\n* Specify the keyword argument ``` skip_header=1 ``` so that the header row is skipped.",
"_____no_output_____"
]
],
[
[
"wines = np.genfromtxt(\"winequality-red.csv\", delimiter=\";\", skip_header=1)\nwines",
"_____no_output_____"
]
],
[
[
"Wines will end up looking the same as if we read it into a list then converted it to an array of ```floats```. NumPy will automatically pick a data type for the elements in an array based on their format.",
"_____no_output_____"
],
[
"## Indexing NumPy Arrays\n\nWe now know how to create arrays, but unless we can retrieve results from them, there isn't a lot we can do with NumPy. We can use array indexing to select individual elements, groups of elements, or entire rows and columns. \n\nOne important thing to keep in mind is that just like Python lists, NumPy is **zero-indexed**, meaning that:\n\n* The index of the first row is 0\n* The index of the first column is 0 \n* If we want to work with the fourth row, we'd use index 3\n* If we want to work with the second row, we'd use index 1, and so on. \n\nWe'll again work with the wines array:\n\n|||||||||||||\n|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|-:|\n|7.4 |0.70\t|0.00\t|1.9\t|0.076\t|11\t|34\t|0.9978\t|3.51\t|0.56\t|9.4\t|5|\n|7.8 |0.88\t|0.00\t|2.6\t|0.098\t|25\t|67\t|0.9968\t|3.20\t|0.68\t|9.8\t|5|\n|7.8 |0.76\t|0.04\t|2.3\t|0.092\t|15\t|54\t|0.9970\t|3.26\t|0.65\t|9.8\t|5|\n|11.2|0.28\t|0.56\t|1.9\t|0.075\t|17\t|60\t|0.9980\t|3.16\t|0.58\t|9.8\t|6|\n|7.4 |0.70\t|0.00\t|1.9\t|0.076\t|11\t|34\t|0.9978\t|3.51\t|0.56\t|9.4\t|5|",
"_____no_output_____"
],
[
"Let's select the element at **row 3** and **column 4**.\n\nWe pass:\n* 2 as the row index\n* 3 as the column index. \n\nThis retrieves the value from the **third row** and **fourth column**",
"_____no_output_____"
]
],
[
[
"wines[2, 3]",
"_____no_output_____"
],
[
"wines[2][3]",
"_____no_output_____"
]
],
[
[
"Since we're working with a 2-dimensional array in NumPy we specify 2 indexes to retrieve an element. \n\n* The first index is the row, or **axis 1**, index\n* The second index is the column, or **axis 2**, index \n\nAny element in wines can be retrieved using 2 indexes.",
"_____no_output_____"
]
],
[
[
"# rows 1, 2, 3 and column 4\nwines[0:3, 3]",
"_____no_output_____"
],
[
"# all rows and column 3\nwines[:, 2]",
"_____no_output_____"
]
],
[
[
"Just like with ```list``` slicing, it's possible to omit the 0 to just retrieve all the elements from the beginning up to element 3:",
"_____no_output_____"
]
],
[
[
"# rows 1, 2, 3 and column 4\nwines[:3, 3]",
"_____no_output_____"
]
],
[
[
"We can select an entire column by specifying that we want all the elements, from the first to the last. We specify this by just using the colon ```:```, with no starting or ending indices. The below code will select the entire fourth column:",
"_____no_output_____"
]
],
[
[
"# all rows and column 4\nwines[:, 3]",
"_____no_output_____"
]
],
[
[
"We selected an entire column above, but we can also extract an entire row:",
"_____no_output_____"
]
],
[
[
"# row 4 and all columns\nwines[3, :]",
"_____no_output_____"
]
],
[
[
"If we take our indexing to the extreme, we can select the entire array using two colons to select all the rows and columns in wines. This is a great party trick, but doesn't have a lot of good applications:",
"_____no_output_____"
]
],
[
[
"wines[:, :]",
"_____no_output_____"
]
],
[
[
"## Assigning Values To NumPy Arrays\nWe can also use indexing to assign values to certain elements in arrays. We can do this by assigning directly to the indexed value:",
"_____no_output_____"
]
],
[
[
"# assign the value of 10 to the 2nd row and 6th column\nprint('Before', wines[1, 4:7])\nwines[1, 5] = 10\nprint('After', wines[1, 4:7])",
"Before [ 0.1 25. 67. ]\nAfter [ 0.1 10. 67. ]\n"
]
],
[
[
"We can do the same for slices. To overwrite an entire column, we can do this:",
"_____no_output_____"
]
],
[
[
"# Overwrites all the values in the eleventh column with 50.\nprint('Before', wines[:, 9:12])\nwines[:, 10] = 50\nprint('After', wines[:, 9:12])",
"Before [[ 0.56 9.4 5. ]\n [ 0.68 9.8 5. ]\n [ 0.65 9.8 5. ]\n ...\n [ 0.75 11. 6. ]\n [ 0.71 10.2 5. ]\n [ 0.66 11. 6. ]]\nAfter [[ 0.56 50. 5. ]\n [ 0.68 50. 5. ]\n [ 0.65 50. 5. ]\n ...\n [ 0.75 50. 6. ]\n [ 0.71 50. 5. ]\n [ 0.66 50. 6. ]]\n"
]
],
[
[
"## 1-Dimensional NumPy Arrays\n\nSo far, we've worked with 2-dimensional arrays, such as wines. However, NumPy is a package for working with multidimensional arrays. \n\nOne of the most common types of multidimensional arrays is the **1-dimensional array**, or **vector**. As you may have noticed above, when we sliced wines, we retrieved a 1-dimensional array. \n\n* A 1-dimensional array only needs a single index to retrieve an element. \n* Each row and column in a 2-dimensional array is a 1-dimensional array. \n\nJust like a list of lists is analogous to a 2-dimensional array, a single list is analogous to a 1-dimensional array. \n\nIf we slice wines and only retrieve the third row, we get a 1-dimensional array:",
"_____no_output_____"
]
],
[
[
"third_wine = wines[3,:]\nthird_wine",
"_____no_output_____"
]
],
[
[
"We can retrieve individual elements from ```third_wine``` using a single index. ",
"_____no_output_____"
]
],
[
[
"# display the second item in third_wine\nthird_wine[1]",
"_____no_output_____"
]
],
[
[
"Most NumPy functions that we've worked with, such as ```numpy.random.rand```, can be used with multidimensional arrays. Here's how we'd use ```numpy.random.rand``` to generate a random vector:",
"_____no_output_____"
]
],
[
[
"np.random.rand(3)",
"_____no_output_____"
]
],
[
[
"Previously, when we called ```np.random.rand```, we passed in a shape for a 2-dimensional array, so the result was a 2-dimensional array. This time, we passed in a shape for a single dimensional array. The shape specifies the number of dimensions, and the size of the array in each dimension. \n\nA shape of ```(10,10)``` will be a 2-dimensional array with **10 rows** and **10 columns**. A shape of ```(10,)``` will be a **1-dimensional** array with **10 elements**.\n\nWhere NumPy gets more complex is when we start to deal with arrays that have more than 2 dimensions.",
"_____no_output_____"
],
[
"## N-Dimensional NumPy Arrays\n\nThis doesn't happen extremely often, but there are cases when you'll want to deal with arrays that have greater than 3 dimensions. One way to think of this is as a list of lists of lists. Let's say we want to store the monthly earnings of a store, but we want to be able to quickly lookup the results for a quarter, and for a year. The earnings for one year might look like this:\n\n``` python\n[500, 505, 490, 810, 450, 678, 234, 897, 430, 560, 1023, 640]\n```\n\nThe store earned \\$500 in January, \\$505 in February, and so on. We can split up these earnings by quarter into a list of lists:",
"_____no_output_____"
]
],
[
[
"year_one = [\n [500,505,490], # 1st quarter\n [810,450,678], # 2nd quarter\n [234,897,430], # 3rd quarter\n [560,1023,640] # 4th quarter\n]",
"_____no_output_____"
]
],
[
[
"We can retrieve the earnings from January by calling ``` year_one[0][0] ```. If we want the results for a whole quarter, we can call ``` year_one[0] ``` or ``` year_one[1] ```. \n\nWe now have a 2-dimensional array, or matrix. But what if we now want to add the results from another year? We have to add a third dimension:",
"_____no_output_____"
]
],
[
[
"earnings = [\n [ # year 1\n [500,505,490], # year 1, 1st quarter\n [810,450,678], # year 1, 2nd quarter\n [234,897,430], # year 1, 3rd quarter\n [560,1023,640] # year 1, 4th quarter\n ],\n [ # year =2\n [600,605,490], # year 2, 1st quarter\n [345,900,1000],# year 2, 2nd quarter\n [780,730,710], # year 2, 3rd quarter\n [670,540,324] # year 2, 4th quarter\n ]\n ]",
"_____no_output_____"
]
],
[
[
"We can retrieve the earnings from January of the first year by calling ``` earnings[0][0][0] ```. \n\nWe now need three indexes to retrieve a single element. A three-dimensional array in NumPy is much the same. In fact, we can convert earnings to an array and then get the earnings for January of the first year:",
"_____no_output_____"
]
],
[
[
"earnings = np.array(earnings)",
"_____no_output_____"
],
[
"# year 1, 1st quarter, 1st month (January)\nearnings[0,0,0] ",
"_____no_output_____"
],
[
"# year 2, 3rd quarter, 1st month (July)\nearnings[1,2,0] ",
"_____no_output_____"
],
[
"# we can also find the shape of the array\nearnings.shape",
"_____no_output_____"
]
],
[
[
"Indexing and slicing work the exact same way with a 3-dimensional array, but now we have an extra axis to pass in. If we wanted to get the earnings for **January of all years**, we could do this:",
"_____no_output_____"
]
],
[
[
"# all years, 1st quarter, 1st month (January)\nearnings[:,0,0]",
"_____no_output_____"
]
],
[
[
"If we wanted to get first quarter earnings from both years, we could do this:",
"_____no_output_____"
]
],
[
[
"# all years, 1st quarter, all months (January, February, March)\nearnings[:,0,:]",
"_____no_output_____"
]
],
[
[
"Adding more dimensions can make it much easier to query your data if it's organized in a certain way. As we go from 3-dimensional arrays to 4-dimensional and larger arrays, the same properties apply, and they can be indexed and sliced in the same ways.",
"_____no_output_____"
],
[
"## NumPy Data Types\n\nAs we mentioned earlier, each NumPy array can store elements of a single data type. For example, wines contains only float values. \n\nNumPy stores values using its own data types, **which are distinct from Python types** like ```float``` and ```str```. \n\nThis is because the core of NumPy is written in a programming language called ```C```, **which stores data differently than the Python data types**. NumPy data types map between Python and C, allowing us to use NumPy arrays without any conversion hitches.\n\nYou can find the data type of a NumPy array by accessing the dtype property:",
"_____no_output_____"
]
],
[
[
"wines.dtype",
"_____no_output_____"
]
],
[
[
"NumPy has several different data types, which mostly map to Python data types, like ```float```, and ```str```. You can find a full listing of NumPy data types [here](https://www.dataquest.io/blog/numpy-tutorial-python/), but here are a few important ones:\n\n* ```float``` -- numeric floating point data.\n* ```int``` -- integer data.\n* ```string``` -- character data.\n* ```object``` -- Python objects.\n\nData types additionally end with a suffix that indicates how many bits of memory they take up. So ```int32``` is a **32 bit integer data type**, and ```float64``` is a **64 bit float data type**.",
"_____no_output_____"
],
[
"### Converting Data Types\n\nYou can use the numpy.ndarray.astype method to convert an array to a different type. The method will actually **copy the array**, and **return a new array with the specified data type**. \n\nFor instance, we can convert wines to the ```int``` data type:",
"_____no_output_____"
]
],
[
[
"# convert wines to the int data type\nwines.astype(int)",
"_____no_output_____"
]
],
[
[
"As you can see above, all of the items in the resulting array are integers. Note that we used the Python ```int``` type instead of a NumPy data type when converting wines. This is because several Python data types, including ```float```, ```int```, and ```string```, can be used with NumPy, and are automatically converted to NumPy data types.\n\nWe can check the name property of the ```dtype``` of the resulting array to see what data type NumPy mapped the resulting array to:",
"_____no_output_____"
]
],
[
[
"# convert to int\nint_wines = wines.astype(int)\n\n# check the data type\nint_wines.dtype.name",
"_____no_output_____"
]
],
[
[
"The array has been converted to a **64-bit integer** data type. This allows for very long integer values, **but takes up more space in memory** than storing the values as 32-bit integers.\n\nIf you want more control over how the array is stored in memory, you can directly create NumPy dtype objects like ```numpy.int32```",
"_____no_output_____"
]
],
[
[
"np.int32",
"_____no_output_____"
]
],
[
[
"You can use these directly to convert between types:",
"_____no_output_____"
]
],
[
[
"# convert to a 64-bit integer\nwines.astype(np.int64)",
"_____no_output_____"
],
[
"# convert to a 32-bit integer\nwines.astype(np.int32)",
"_____no_output_____"
],
[
"# convert to a 16-bit integer\nwines.astype(np.int16)",
"_____no_output_____"
],
[
"# convert to a 8-bit integer\nwines.astype(np.int8)",
"_____no_output_____"
]
],
[
[
"## NumPy Array Operations\n\nNumPy makes it simple to perform mathematical operations on arrays. This is one of the primary advantages of NumPy, and makes it quite easy to do computations.\n\n### Single Array Math\nIf you do any of the basic mathematical operations ```/```, ```*```, ```-```, ```+```, ```^``` with an array and a value, it will apply the operation to each of the elements in the array.\n\nLet's say we want to add 10 points to each quality score because we're feeling generous. Here's how we'd do that:",
"_____no_output_____"
]
],
[
[
"# add 10 points to the quality score\nwines[:,-1] + 10",
"_____no_output_____"
]
],
[
[
"*Note: that the above operation won't change the wines array -- it will return a new 1-dimensional array where 10 has been added to each element in the quality column of wines.*\n\nIf we instead did ```+=```, we'd modify the array in place:",
"_____no_output_____"
]
],
[
[
"print('Before', wines[:,11])\n\n# modify the data in place\nwines[:,11] += 10\n\nprint('After', wines[:,11])",
"Before [5. 5. 5. ... 6. 5. 6.]\nAfter [15. 15. 15. ... 16. 15. 16.]\n"
]
],
[
[
"All the other operations work the same way. For example, if we want to multiply each of the quality score by 2, we could do it like this:",
"_____no_output_____"
]
],
[
[
"# multiply the quality score by 2\nwines[:,11] * 2",
"_____no_output_____"
]
],
[
[
"### Multiple Array Math\n\nIt's also possible to do mathematical operations between arrays. This will apply the operation to pairs of elements. For example, if we add the quality column to itself, here's what we get:",
"_____no_output_____"
]
],
[
[
"# add the quality column to itself\nwines[:,11] + wines[:,11]",
"_____no_output_____"
]
],
[
[
"Note that this is equivalent to ```wines[:,11] * 2``` -- this is because NumPy adds each pair of elements. The first element in the first array is added to the first element in the second array, the second to the second, and so on.",
"_____no_output_____"
]
],
[
[
"# add the quality column to itself\nwines[:,11] * 2",
"_____no_output_____"
]
],
[
[
"We can also use this to multiply arrays. Let's say we want to pick a wine that maximizes alcohol content and quality. We'd multiply alcohol by quality, and select the wine with the highest score:",
"_____no_output_____"
]
],
[
[
"# multiply alcohol content by quality\nalcohol_by_quality = wines[:,10] * wines[:,11]\nprint(alcohol_by_quality)",
"[750. 750. 750. ... 800. 750. 800.]\n"
],
[
"alcohol_by_quality.sort()\nprint(alcohol_by_quality, alcohol_by_quality[-1])",
"[650. 650. 650. ... 900. 900. 900.] 900.0\n"
]
],
[
[
"All of the common operations ```/```, ```*```, ```-```, ```+```, ```^``` will work between arrays.",
"_____no_output_____"
],
[
"## NumPy Array Methods\n\nIn addition to the common mathematical operations, NumPy also has several methods that you can use for more complex calculations on arrays. An example of this is the ```numpy.ndarray.sum``` method. This finds the sum of all the elements in an array by default:",
"_____no_output_____"
]
],
[
[
"# find the sum of all rows and the quality column\ntotal = 0\nfor row in wines:\n total += row[11]\nprint(total)",
"25002.0\n"
],
[
"# find the sum of all rows and the quality column\nwines[:,11].sum(axis=0)",
"_____no_output_____"
],
[
"# find the sum of the rows 1, 2, and 3 across all columns\ntotals = []\nfor i in range(3):\n total = 0\n for col in wines[i,:]:\n total += col\n totals.append(total)\nprint(totals)",
"[125.1438, 158.2548, 149.899]\n"
],
[
"# find the sum of the rows 1, 2, and 3 across all columns\nwines[0:3,:].sum(axis=1)",
"_____no_output_____"
]
],
[
[
"We can pass the ```axis``` keyword argument into the sum method to find sums over an axis. \n\nIf we call sum across the wines matrix, and pass in ```axis=0```, we'll find the sums over the first axis of the array. This will give us the **sum of all the values in every column**. \n\nThis may seem backwards that the sums over the first axis would give us the sum of each column, but one way to think about this is that **the specified axis is the one \"going away\"**. \n\nSo if we specify ```axis=0```, we want the **rows to go away**, and we want to find **the sums for each of the remaining axes across each row**:",
"_____no_output_____"
]
],
[
[
"# sum each column for all rows\ntotals = [0] * len(wines[0])\nfor i, total in enumerate(totals):\n for row_val in wines[:,i]:\n total += row_val\n totals[i] = total\nprint(totals)",
"[13303.100000000046, 843.9850000000005, 433.2899999999982, 4059.550000000003, 139.8589999999996, 25369.0, 74302.0, 1593.7979399999986, 5294.470000000001, 1052.3800000000006, 79950.0, 25002.0]\n"
],
[
"# sum each column for all rows\nwines.sum(axis=0)",
"_____no_output_____"
]
],
[
[
"We can verify that we did the sum correctly by checking the shape. The shape should be 12, corresponding to the number of columns:",
"_____no_output_____"
]
],
[
[
"wines.sum(axis=0).shape",
"_____no_output_____"
]
],
[
[
"If we pass in axis=1, we'll find the sums over the second axis of the array. This will give us the sum of each row:",
"_____no_output_____"
]
],
[
[
"# sum each row for all columns\ntotals = [0] * len(wines)\nfor i, total in enumerate(totals):\n for col_val in wines[i,:]:\n total += col_val\n totals[i] = total\nprint(totals[0:3], '...', totals[-3:])",
"[125.1438, 158.2548, 149.899] ... [149.48174, 155.01547, 141.49249]\n"
],
[
"# sum each row for all columns\nwines.sum(axis=1)",
"_____no_output_____"
],
[
"wines.sum(axis=1).shape",
"_____no_output_____"
]
],
[
[
"There are several other methods that behave like the sum method, including:\n\n* ```numpy.ndarray.mean``` — finds the mean of an array.\n* ```numpy.ndarray.std``` — finds the standard deviation of an array.\n* ```numpy.ndarray.min``` — finds the minimum value in an array.\n* ```numpy.ndarray.max``` — finds the maximum value in an array.\n\nYou can find a full list of array methods [here](http://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html).",
"_____no_output_____"
],
[
"## NumPy Array Comparisons\n\nNumPy makes it possible to test to see if rows match certain values using mathematical comparison operations like ```<```, ```>```, ```>=```, ```<=```, and ```==```. For example, if we want to see which wines have a quality rating higher than 5, we can do this:",
"_____no_output_____"
]
],
[
[
"# return True for all rows in the Quality column that are greater than 5\nwines[:,11] > 5",
"_____no_output_____"
]
],
[
[
"We get a Boolean array that tells us which of the wines have a quality rating greater than 5. We can do something similar with the other operators. For instance, we can see if any wines have a quality rating equal to 10:",
"_____no_output_____"
]
],
[
[
"# return True for all rows that have a Quality rating of 10\nwines[:,11] == 10",
"_____no_output_____"
]
],
[
[
"### Subsetting\n\nOne of the powerful things we can do with a Boolean array and a NumPy array is select only certain rows or columns in the NumPy array. For example, the below code will only select rows in wines where the quality is over 7:",
"_____no_output_____"
]
],
[
[
"# create a boolean array for wines with quality greater than 15\nhigh_quality = wines[:,11] > 15\nprint(len(high_quality), high_quality)",
"1599 [False False False ... True False True]\n"
],
[
"# use boolean indexing to find high quality wines\nhigh_quality_wines = wines[high_quality,:]\nprint(len(high_quality_wines), high_quality_wines)",
"855 [[1.12e+01 2.80e-01 5.60e-01 ... 5.80e-01 5.00e+01 1.60e+01]\n [7.30e+00 6.50e-01 0.00e+00 ... 4.70e-01 5.00e+01 1.70e+01]\n [7.80e+00 5.80e-01 2.00e-02 ... 5.70e-01 5.00e+01 1.70e+01]\n ...\n [5.90e+00 5.50e-01 1.00e-01 ... 7.60e-01 5.00e+01 1.60e+01]\n [6.30e+00 5.10e-01 1.30e-01 ... 7.50e-01 5.00e+01 1.60e+01]\n [6.00e+00 3.10e-01 4.70e-01 ... 6.60e-01 5.00e+01 1.60e+01]]\n"
]
],
[
[
"We select only the rows where ```high_quality``` contains a ```True``` value, and all of the columns. This subsetting makes it simple to filter arrays for certain criteria. \n\nFor example, we can look for wines with a lot of alcohol and high quality. In order to specify multiple conditions, we have to place each condition in **parentheses** ```(...)```, and separate conditions with an **ampersand** ```&```:",
"_____no_output_____"
]
],
[
[
"# create a boolean array for high alcohol content and high quality\nhigh_alcohol_and_quality = (wines[:,11] > 7) & (wines[:,10] > 10)\nprint(high_alcohol_and_quality)\n\n# use boolean indexing to select out the wines\nwines[high_alcohol_and_quality,:]",
"[ True True True ... True True True]\n"
]
],
[
[
"We can combine subsetting and assignment to overwrite certain values in an array:",
"_____no_output_____"
]
],
[
[
"high_alcohol_and_quality = (wines[:,10] > 10) & (wines[:,11] > 7)\nwines[high_alcohol_and_quality,10:] = 20",
"_____no_output_____"
]
],
[
[
"## Reshaping NumPy Arrays\n\nWe can change the shape of arrays while still preserving all of their elements. This often can make it easier to access array elements. The simplest reshaping is to flip the axes, so rows become columns, and vice versa. We can accomplish this with the ```numpy.transpose``` function:",
"_____no_output_____"
]
],
[
[
"np.transpose(wines).shape",
"_____no_output_____"
]
],
[
[
"We can use the ```numpy.ravel``` function to turn an array into a one-dimensional representation. It will essentially flatten an array into a long sequence of values:",
"_____no_output_____"
]
],
[
[
"wines.ravel()",
"_____no_output_____"
]
],
[
[
"Here's an example where we can see the ordering of ```numpy.ravel```:",
"_____no_output_____"
]
],
[
[
"array_one = np.array(\n [\n [1, 2, 3, 4], \n [5, 6, 7, 8]\n ]\n)\n\narray_one.ravel()",
"_____no_output_____"
]
],
[
[
"Finally, we can use the numpy.reshape function to reshape an array to a certain shape we specify. The below code will turn the second row of wines into a 2-dimensional array with 2 rows and 6 columns:",
"_____no_output_____"
]
],
[
[
"# print the current shape of the 2nd row and all columns\nwines[1,:].shape",
"_____no_output_____"
],
[
"# reshape the 2nd row to a 2 by 6 matrix\nwines[1,:].reshape((2,6))",
"_____no_output_____"
]
],
[
[
"## Combining NumPy Arrays\n\nWith NumPy, it's very common to combine multiple arrays into a single unified array. We can use ```numpy.vstack``` to vertically stack multiple arrays. \n\nThink of it like the second arrays's items being added as new rows to the first array. We can read in the ```winequality-white.csv``` dataset that contains information on the quality of white wines, then combine it with our existing dataset, wines, which contains information on red wines.\n\nIn the below code, we:\n\n* Read in ```winequality-white.csv```.\n* Display the shape of white_wines.",
"_____no_output_____"
]
],
[
[
"white_wines = np.genfromtxt(\"winequality-white.csv\", delimiter=\";\", skip_header=1)\nwhite_wines.shape",
"_____no_output_____"
]
],
[
[
"As you can see, we have attributes for 4898 wines. Now that we have the white wines data, we can combine all the wine data.\n\nIn the below code, we:\n\n* Use the ```vstack``` function to combine wines and white_wines.\n* Display the shape of the result.",
"_____no_output_____"
]
],
[
[
"all_wines = np.vstack((wines, white_wines))\nall_wines.shape",
"_____no_output_____"
]
],
[
[
"As you can see, the result has 6497 rows, which is the sum of the number of rows in wines and the number of rows in red_wines.\n\nIf we want to combine arrays horizontally, where the number of rows stay constant, but the columns are joined, then we can use the ```numpy.hstack``` function. The arrays we combine need to have the same number of rows for this to work.\n\nFinally, we can use ```numpy.concatenate``` as a general purpose version of ```hstack``` and ```vstack```. If we want to concatenate two arrays, we pass them into concatenate, then specify the axis keyword argument that we want to concatenate along. \n\n* Concatenating along the first axis is similar to ```vstack```\n* Concatenating along the second axis is similar to ```hstack```:",
"_____no_output_____"
]
],
[
[
"x = np.concatenate((wines, white_wines), axis=0)\nprint(x.shape, x)",
"(6497, 12) [[7.40e+00 7.00e-01 0.00e+00 ... 5.60e-01 5.00e+01 1.50e+01]\n [7.80e+00 8.80e-01 0.00e+00 ... 6.80e-01 5.00e+01 1.50e+01]\n [7.80e+00 7.60e-01 4.00e-02 ... 6.50e-01 5.00e+01 1.50e+01]\n ...\n [6.50e+00 2.40e-01 1.90e-01 ... 4.60e-01 9.40e+00 6.00e+00]\n [5.50e+00 2.90e-01 3.00e-01 ... 3.80e-01 1.28e+01 7.00e+00]\n [6.00e+00 2.10e-01 3.80e-01 ... 3.20e-01 1.18e+01 6.00e+00]]\n"
]
],
[
[
"## Broadcasting\n\nUnless the arrays that you're operating on are the exact same size, it's not possible to do elementwise operations. In cases like this, NumPy performs broadcasting to try to match up elements. Essentially, broadcasting involves a few steps:\n\n* The last dimension of each array is compared.\n * If the dimension lengths are equal, or one of the dimensions is of length 1, then we keep going.\n * If the dimension lengths aren't equal, and none of the dimensions have length 1, then there's an error.\n* Continue checking dimensions until the shortest array is out of dimensions.\n\nFor example, the following two shapes are compatible:\n\n``` python\nA: (50,3)\nB (3,)\n```\n\nThis is because the length of the trailing dimension of array A is 3, and the length of the trailing dimension of array B is 3. They're equal, so that dimension is okay. Array B is then out of elements, so we're okay, and the arrays are compatible for mathematical operations.\n\nThe following two shapes are also compatible:\n\n``` python\nA: (1,2)\nB (50,2)\n```\n\nThe last dimension matches, and A is of length 1 in the first dimension.\n\nThese two arrays don't match:\n\n``` python\nA: (50,50)\nB: (49,49)\n```\n\nThe lengths of the dimensions aren't equal, and neither array has either dimension length equal to 1.\n\nThere's a detailed explanation of broadcasting [here](http://docs.scipy.org/doc/numpy/user/basics.broadcasting.html), but we'll go through a few examples to illustrate the principle:\n",
"_____no_output_____"
]
],
[
[
"wines * np.array([1,2])",
"_____no_output_____"
]
],
[
[
"The above example didn't work because the two arrays don't have a matching trailing dimension. Here's an example where the last dimension does match:",
"_____no_output_____"
]
],
[
[
"array_one = np.array(\n [\n [1,2],\n [3,4]\n ]\n)\narray_two = np.array([4,5])\n\narray_one + array_two",
"_____no_output_____"
]
],
[
[
"As you can see, array_two has been broadcasted across each row of array_one. Here's an example with our wines data:",
"_____no_output_____"
]
],
[
[
"rand_array = np.random.rand(12)\nwines + rand_array",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b73f201a84bbcfa1f873c57d471afc576112aa | 13,678 | ipynb | Jupyter Notebook | doc/source/cookbook/fits_xray_images.ipynb | Kiradorn/yt | d4d833bd773413e776b2c5b0f771e34857c27f74 | [
"BSD-3-Clause-Clear"
] | 360 | 2017-04-24T05:06:04.000Z | 2022-03-31T10:47:07.000Z | doc/source/cookbook/fits_xray_images.ipynb | Kiradorn/yt | d4d833bd773413e776b2c5b0f771e34857c27f74 | [
"BSD-3-Clause-Clear"
] | 2,077 | 2017-04-20T20:36:07.000Z | 2022-03-31T16:39:43.000Z | doc/source/cookbook/fits_xray_images.ipynb | Kiradorn/yt | d4d833bd773413e776b2c5b0f771e34857c27f74 | [
"BSD-3-Clause-Clear"
] | 257 | 2017-04-19T20:52:28.000Z | 2022-03-29T12:23:52.000Z | 27.356 | 485 | 0.542258 | [
[
[
"%matplotlib inline\nimport numpy as np\n\nimport yt",
"_____no_output_____"
]
],
[
[
"This notebook shows how to use yt to make plots and examine FITS X-ray images and events files. ",
"_____no_output_____"
],
[
"## Sloshing, Shocks, and Bubbles in Abell 2052",
"_____no_output_____"
],
[
"This example uses data provided by [Scott Randall](http://hea-www.cfa.harvard.edu/~srandall/), presented originally in [Blanton, E.L., Randall, S.W., Clarke, T.E., et al. 2011, ApJ, 737, 99](https://ui.adsabs.harvard.edu/abs/2011ApJ...737...99B). They consist of two files, a \"flux map\" in counts/s/pixel between 0.3 and 2 keV, and a spectroscopic temperature map in keV. ",
"_____no_output_____"
]
],
[
[
"ds = yt.load(\n \"xray_fits/A2052_merged_0.3-2_match-core_tmap_bgecorr.fits\",\n auxiliary_files=[\"xray_fits/A2052_core_tmap_b1_m2000_.fits\"],\n)",
"_____no_output_____"
]
],
[
[
"Since the flux and projected temperature images are in two different files, we had to use one of them (in this case the \"flux\" file) as a master file, and pass in the \"temperature\" file with the `auxiliary_files` keyword to `load`. ",
"_____no_output_____"
],
[
"Next, let's derive some new fields for the number of counts, the \"pseudo-pressure\", and the \"pseudo-entropy\":",
"_____no_output_____"
]
],
[
[
"def _counts(field, data):\n exposure_time = data.get_field_parameter(\"exposure_time\")\n return data[\"fits\", \"flux\"] * data[\"fits\", \"pixel\"] * exposure_time\n\n\nds.add_field(\n (\"gas\", \"counts\"),\n function=_counts,\n sampling_type=\"cell\",\n units=\"counts\",\n take_log=False,\n)\n\n\ndef _pp(field, data):\n return np.sqrt(data[\"gas\", \"counts\"]) * data[\"fits\", \"projected_temperature\"]\n\n\nds.add_field(\n (\"gas\", \"pseudo_pressure\"),\n function=_pp,\n sampling_type=\"cell\",\n units=\"sqrt(counts)*keV\",\n take_log=False,\n)\n\n\ndef _pe(field, data):\n return data[\"fits\", \"projected_temperature\"] * data[\"gas\", \"counts\"] ** (-1.0 / 3.0)\n\n\nds.add_field(\n (\"gas\", \"pseudo_entropy\"),\n function=_pe,\n sampling_type=\"cell\",\n units=\"keV*(counts)**(-1/3)\",\n take_log=False,\n)",
"_____no_output_____"
]
],
[
[
"Here, we're deriving a \"counts\" field from the \"flux\" field by passing it a `field_parameter` for the exposure time of the time and multiplying by the pixel scale. Second, we use the fact that the surface brightness is strongly dependent on density ($S_X \\propto \\rho^2$) to use the counts in each pixel as a \"stand-in\". Next, we'll grab the exposure time from the primary FITS header of the flux file and create a `YTQuantity` from it, to be used as a `field_parameter`:",
"_____no_output_____"
]
],
[
[
"exposure_time = ds.quan(ds.primary_header[\"exposure\"], \"s\")",
"_____no_output_____"
]
],
[
[
"Now, we can make the `SlicePlot` object of the fields we want, passing in the `exposure_time` as a `field_parameter`. We'll also set the width of the image to 250 pixels.",
"_____no_output_____"
]
],
[
[
"slc = yt.SlicePlot(\n ds,\n \"z\",\n [\n (\"fits\", \"flux\"),\n (\"fits\", \"projected_temperature\"),\n (\"gas\", \"pseudo_pressure\"),\n (\"gas\", \"pseudo_entropy\"),\n ],\n origin=\"native\",\n field_parameters={\"exposure_time\": exposure_time},\n)\nslc.set_log((\"fits\", \"flux\"), True)\nslc.set_log((\"gas\", \"pseudo_pressure\"), False)\nslc.set_log((\"gas\", \"pseudo_entropy\"), False)\nslc.set_width(250.0)\nslc.show()",
"_____no_output_____"
]
],
[
[
"To add the celestial coordinates to the image, we can use `PlotWindowWCS`, if you have a recent version of AstroPy (>= 1.3) installed:",
"_____no_output_____"
]
],
[
[
"from yt.frontends.fits.misc import PlotWindowWCS\n\nwcs_slc = PlotWindowWCS(slc)\nwcs_slc.show()",
"_____no_output_____"
]
],
[
[
"We can make use of yt's facilities for profile plotting as well.",
"_____no_output_____"
]
],
[
[
"v, c = ds.find_max((\"fits\", \"flux\")) # Find the maximum flux and its center\nmy_sphere = ds.sphere(c, (100.0, \"code_length\")) # Radius of 150 pixels\nmy_sphere.set_field_parameter(\"exposure_time\", exposure_time)",
"_____no_output_____"
]
],
[
[
"Such as a radial profile plot:",
"_____no_output_____"
]
],
[
[
"radial_profile = yt.ProfilePlot(\n my_sphere,\n \"radius\",\n [\"counts\", \"pseudo_pressure\", \"pseudo_entropy\"],\n n_bins=30,\n weight_field=\"ones\",\n)\nradial_profile.set_log(\"counts\", True)\nradial_profile.set_log(\"pseudo_pressure\", True)\nradial_profile.set_log(\"pseudo_entropy\", True)\nradial_profile.set_xlim(3, 100.0)\nradial_profile.show()",
"_____no_output_____"
]
],
[
[
"Or a phase plot:",
"_____no_output_____"
]
],
[
[
"phase_plot = yt.PhasePlot(\n my_sphere, \"pseudo_pressure\", \"pseudo_entropy\", [\"counts\"], weight_field=None\n)\nphase_plot.show()",
"_____no_output_____"
]
],
[
[
"Finally, we can also take an existing [ds9](http://ds9.si.edu/site/Home.html) region and use it to create a \"cut region\", using `ds9_region` (the [pyregion](https://pyregion.readthedocs.io) package needs to be installed for this):",
"_____no_output_____"
]
],
[
[
"from yt.frontends.fits.misc import ds9_region\n\nreg_file = [\n \"# Region file format: DS9 version 4.1\\n\",\n \"global color=green dashlist=8 3 width=3 include=1 source=1 fk5\\n\",\n 'circle(15:16:44.817,+7:01:19.62,34.6256\")',\n]\nf = open(\"circle.reg\", \"w\")\nf.writelines(reg_file)\nf.close()\ncircle_reg = ds9_region(\n ds, \"circle.reg\", field_parameters={\"exposure_time\": exposure_time}\n)",
"_____no_output_____"
]
],
[
[
"This region may now be used to compute derived quantities:",
"_____no_output_____"
]
],
[
[
"print(\n circle_reg.quantities.weighted_average_quantity(\"projected_temperature\", \"counts\")\n)",
"_____no_output_____"
]
],
[
[
"Or used in projections:",
"_____no_output_____"
]
],
[
[
"prj = yt.ProjectionPlot(\n ds,\n \"z\",\n [\n (\"fits\", \"flux\"),\n (\"fits\", \"projected_temperature\"),\n (\"gas\", \"pseudo_pressure\"),\n (\"gas\", \"pseudo_entropy\"),\n ],\n origin=\"native\",\n field_parameters={\"exposure_time\": exposure_time},\n data_source=circle_reg,\n method=\"sum\",\n)\nprj.set_log((\"fits\", \"flux\"), True)\nprj.set_log((\"gas\", \"pseudo_pressure\"), False)\nprj.set_log((\"gas\", \"pseudo_entropy\"), False)\nprj.set_width(250.0)\nprj.show()",
"_____no_output_____"
]
],
[
[
"## The Bullet Cluster",
"_____no_output_____"
],
[
"This example uses an events table file from a ~100 ks exposure of the \"Bullet Cluster\" from the [Chandra Data Archive](http://cxc.harvard.edu/cda/). In this case, the individual photon events are treated as particle fields in yt. However, you can make images of the object in different energy bands using the `setup_counts_fields` function. ",
"_____no_output_____"
]
],
[
[
"from yt.frontends.fits.api import setup_counts_fields",
"_____no_output_____"
]
],
[
[
"`load` will handle the events file as FITS image files, and will set up a grid using the WCS information in the file. Optionally, the events may be reblocked to a new resolution. by setting the `\"reblock\"` parameter in the `parameters` dictionary in `load`. `\"reblock\"` must be a power of 2. ",
"_____no_output_____"
]
],
[
[
"ds2 = yt.load(\"xray_fits/acisf05356N003_evt2.fits.gz\", parameters={\"reblock\": 2})",
"_____no_output_____"
]
],
[
[
"`setup_counts_fields` will take a list of energy bounds (emin, emax) in keV and create a new field from each where the photons in that energy range will be deposited onto the image grid. ",
"_____no_output_____"
]
],
[
[
"ebounds = [(0.1, 2.0), (2.0, 5.0)]\nsetup_counts_fields(ds2, ebounds)",
"_____no_output_____"
]
],
[
[
"The \"x\", \"y\", \"energy\", and \"time\" fields in the events table are loaded as particle fields. Each one has a name given by \"event\\_\" plus the name of the field:",
"_____no_output_____"
]
],
[
[
"dd = ds2.all_data()\nprint(dd[\"io\", \"event_x\"])\nprint(dd[\"io\", \"event_y\"])",
"_____no_output_____"
]
],
[
[
"Now, we'll make a plot of the two counts fields we made, and pan and zoom to the bullet:",
"_____no_output_____"
]
],
[
[
"slc = yt.SlicePlot(\n ds2, \"z\", [(\"gas\", \"counts_0.1-2.0\"), (\"gas\", \"counts_2.0-5.0\")], origin=\"native\"\n)\nslc.pan((100.0, 100.0))\nslc.set_width(500.0)\nslc.show()",
"_____no_output_____"
]
],
[
[
"The counts fields can take the field parameter `\"sigma\"` and use [AstroPy's convolution routines](https://astropy.readthedocs.io/en/latest/convolution/) to smooth the data with a Gaussian:",
"_____no_output_____"
]
],
[
[
"slc = yt.SlicePlot(\n ds2,\n \"z\",\n [(\"gas\", \"counts_0.1-2.0\"), (\"gas\", \"counts_2.0-5.0\")],\n origin=\"native\",\n field_parameters={\"sigma\": 2.0},\n) # This value is in pixel scale\nslc.pan((100.0, 100.0))\nslc.set_width(500.0)\nslc.set_zlim((\"gas\", \"counts_0.1-2.0\"), 0.01, 100.0)\nslc.set_zlim((\"gas\", \"counts_2.0-5.0\"), 0.01, 50.0)\nslc.show()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b740a02e8eb2822d5ccece30f2ba150a274d85 | 110,604 | ipynb | Jupyter Notebook | examples/Example_OLR_Tracking_satellite/Example_OLR_Tracking_satellite.ipynb | rcjackson/tobac | 9f3b9812e9a13a26373e42d356f7d571366bb967 | [
"BSD-3-Clause"
] | 36 | 2018-11-12T10:42:22.000Z | 2022-03-08T04:29:58.000Z | examples/Example_OLR_Tracking_satellite/Example_OLR_Tracking_satellite.ipynb | rcjackson/tobac | 9f3b9812e9a13a26373e42d356f7d571366bb967 | [
"BSD-3-Clause"
] | 71 | 2018-12-04T13:11:54.000Z | 2022-03-30T23:15:26.000Z | examples/Example_OLR_Tracking_satellite/Example_OLR_Tracking_satellite.ipynb | rcjackson/tobac | 9f3b9812e9a13a26373e42d356f7d571366bb967 | [
"BSD-3-Clause"
] | 28 | 2018-11-19T07:51:02.000Z | 2022-02-17T16:26:40.000Z | 197.15508 | 87,460 | 0.904832 | [
[
[
"tobac example: Tracking deep convection based on OLR from geostationary satellite retrievals\n==\n\nThis example notebook demonstrates the use of tobac to track isolated deep convective clouds based on outgoing longwave radiation (OLR) calculated based on a combination of two different channels of the GOES-13 imaging instrument.\n\nThe data used in this example is downloaded from \"zenodo link\" automatically as part of the notebooks (This only has to be done once for all the tobac example notebooks).",
"_____no_output_____"
]
],
[
[
"# Import libraries:\nimport iris\nimport numpy as np\nimport pandas as pd\nimport os\nimport matplotlib.pyplot as plt\nimport iris.plot as iplt\nimport iris.quickplot as qplt\nimport zipfile\nfrom six.moves import urllib\nfrom glob import glob\n%matplotlib inline",
"_____no_output_____"
],
[
"# Import tobac itself:\nimport tobac",
"_____no_output_____"
],
[
"# Disable a few warnings:\nimport warnings\nwarnings.filterwarnings('ignore', category=UserWarning, append=True)\nwarnings.filterwarnings('ignore', category=RuntimeWarning, append=True)\nwarnings.filterwarnings('ignore', category=FutureWarning, append=True)\nwarnings.filterwarnings('ignore',category=pd.io.pytables.PerformanceWarning)\n",
"_____no_output_____"
]
],
[
[
"**Download example data:** \nThis has to be done only once for all tobac examples.",
"_____no_output_____"
]
],
[
[
"data_out='../'",
"_____no_output_____"
],
[
"# # Download the data: This only has to be done once for all tobac examples and can take a while\n# file_path='https://zenodo.org/record/3195910/files/climate-processes/tobac_example_data-v1.0.1.zip'\n# tempfile='temp.zip'\n# print('start downloading data')\n# request=urllib.request.urlretrieve(file_path,tempfile)\n# print('start extracting data')\n# zf = zipfile.ZipFile(tempfile)\n# zf.extractall(data_out)\n# print('example data saved in')",
"_____no_output_____"
]
],
[
[
"**Load data:**",
"_____no_output_____"
]
],
[
[
"data_file=os.path.join(data_out,'*','data','Example_input_OLR_satellite.nc')\ndata_file = glob(data_file)[0]",
"_____no_output_____"
],
[
"print(data_file)",
"../climate-processes-tobac_example_data-b3e69ee/data/Example_input_OLR_satellite.nc\n"
],
[
"# Load Data from downloaded file:\nOLR=iris.load_cube(data_file,'OLR')",
"_____no_output_____"
],
[
"# Display information about the input data cube:\ndisplay(OLR)",
"_____no_output_____"
],
[
"#Set up directory to save output and plots:\nsavedir='Save'\nif not os.path.exists(savedir):\n os.makedirs(savedir)\nplot_dir=\"Plot\"\nif not os.path.exists(plot_dir):\n os.makedirs(plot_dir)",
"_____no_output_____"
]
],
[
[
"**Feature identification:** \nIdentify features based on OLR field and a set of threshold values",
"_____no_output_____"
]
],
[
[
"# Determine temporal and spatial sampling of the input data:\ndxy,dt=tobac.get_spacings(OLR,grid_spacing=4000)",
"_____no_output_____"
],
[
"# Keyword arguments for the feature detection step\nparameters_features={}\nparameters_features['position_threshold']='weighted_diff'\nparameters_features['sigma_threshold']=0.5\nparameters_features['min_num']=4\nparameters_features['target']='minimum'\nparameters_features['threshold']=[250,225,200,175,150]",
"_____no_output_____"
],
[
"# Feature detection and save results to file:\nprint('starting feature detection')\nFeatures=tobac.feature_detection_multithreshold(OLR,dxy,**parameters_features)\nFeatures.to_hdf(os.path.join(savedir,'Features.h5'),'table')\nprint('feature detection performed and saved')\n",
"starting feature detection\nfeature detection performed and saved\n"
]
],
[
[
"**Segmentation:** \nSegmentation is performed based on the OLR field and a threshold value to determine the cloud areas.",
"_____no_output_____"
]
],
[
[
"# Keyword arguments for the segmentation step:\nparameters_segmentation={}\nparameters_segmentation['target']='minimum'\nparameters_segmentation['method']='watershed'\nparameters_segmentation['threshold']=250",
"_____no_output_____"
],
[
"# Perform segmentation and save results to files:\nMask_OLR,Features_OLR=tobac.segmentation_2D(Features,OLR,dxy,**parameters_segmentation)\nprint('segmentation OLR performed, start saving results to files')\niris.save([Mask_OLR],os.path.join(savedir,'Mask_Segmentation_OLR.nc'),zlib=True,complevel=4) \nFeatures_OLR.to_hdf(os.path.join(savedir,'Features_OLR.h5'),'table')\nprint('segmentation OLR performed and saved')",
"segmentation OLR performed, start saving results to files\nsegmentation OLR performed and saved\n"
]
],
[
[
"**Trajectory linking:** \nThe detected features are linked into cloud trajectories using the trackpy library (http://soft-matter.github.io/trackpy). This takes the feature positions determined in the feature detection step into account but does not include information on the shape of the identified objects.",
"_____no_output_____"
]
],
[
[
"# keyword arguments for linking step\nparameters_linking={}\nparameters_linking['v_max']=20\nparameters_linking['stubs']=2\nparameters_linking['order']=1\nparameters_linking['extrapolate']=1\nparameters_linking['memory']=0\nparameters_linking['adaptive_stop']=0.2\nparameters_linking['adaptive_step']=0.95\nparameters_linking['subnetwork_size']=100\nparameters_linking['method_linking']= 'predict'",
"_____no_output_____"
],
[
"# Perform linking and save results to file:\nTrack=tobac.linking_trackpy(Features,OLR,dt=dt,dxy=dxy,**parameters_linking)\nTrack.to_hdf(os.path.join(savedir,'Track.h5'),'table')",
"Frame 53: 19 trajectories present.\n"
]
],
[
[
"**Visualisation:** ",
"_____no_output_____"
]
],
[
[
"# Set extent of maps created in the following cells:\naxis_extent=[-95,-89,28,32] ",
"_____no_output_____"
],
[
"# Plot map with all individual tracks:\nimport cartopy.crs as ccrs\nfig_map,ax_map=plt.subplots(figsize=(10,10),subplot_kw={'projection': ccrs.PlateCarree()})\nax_map=tobac.map_tracks(Track,axis_extent=axis_extent,axes=ax_map)",
"_____no_output_____"
],
[
"# Create animation of tracked clouds and outlines with OLR as a background field\nanimation_test_tobac=tobac.animation_mask_field(Track,Features,OLR,Mask_OLR,\n axis_extent=axis_extent,#figsize=figsize,orientation_colorbar='horizontal',pad_colorbar=0.2,\n vmin=80,vmax=330,cmap='Blues_r',\n plot_outline=True,plot_marker=True,marker_track='x',plot_number=True,plot_features=True)",
"_____no_output_____"
],
[
"# Display animation:\nfrom IPython.display import HTML, Image, display\nHTML(animation_test_tobac.to_html5_video())",
"_____no_output_____"
],
[
"# # Save animation to file:\n# savefile_animation=os.path.join(plot_dir,'Animation.mp4')\n# animation_test_tobac.save(savefile_animation,dpi=200)\n# print(f'animation saved to {savefile_animation}')",
"_____no_output_____"
],
[
"# Lifetimes of tracked clouds:\nfig_lifetime,ax_lifetime=plt.subplots()\ntobac.plot_lifetime_histogram_bar(Track,axes=ax_lifetime,bin_edges=np.arange(0,200,20),density=False,width_bar=10)\nax_lifetime.set_xlabel('lifetime (min)')\nax_lifetime.set_ylabel('counts')\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b7529b16bea6dcc1f1f2cf9aaa2eacd951ed13 | 260,994 | ipynb | Jupyter Notebook | experiments/tl_1v2/oracle.run1.framed-oracle.run2.framed/trials/27/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tl_1v2/oracle.run1.framed-oracle.run2.framed/trials/27/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | experiments/tl_1v2/oracle.run1.framed-oracle.run2.framed/trials/27/trial.ipynb | stevester94/csc500-notebooks | 4c1b04c537fe233a75bed82913d9d84985a89177 | [
"MIT"
] | null | null | null | 97.132118 | 75,892 | 0.783597 | [
[
[
"# Transfer Learning Template",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom torch.utils.data import DataLoader\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Allowed Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"n_shot\",\n \"n_query\",\n \"n_way\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_net\",\n \"datasets\",\n \"torch_default_dtype\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"x_shape\",\n}",
"_____no_output_____"
],
[
"from steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\nfrom steves_utils.ORACLE.utils_v2 import (\n ALL_DISTANCES_FEET_NARROWED,\n ALL_RUNS,\n ALL_SERIAL_NUMBERS,\n)\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\nstandalone_parameters[\"n_way\"] = 8\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 50\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"source_loss\"\n\nstandalone_parameters[\"datasets\"] = [\n {\n \"labels\": ALL_SERIAL_NUMBERS,\n \"domains\": ALL_DISTANCES_FEET_NARROWED,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"minus_two\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE_\"\n },\n {\n \"labels\": ALL_NODES,\n \"domains\": ALL_DAYS,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_power\", \"times_zero\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\"\n } \n]\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n\n\n\n",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"tl_1v2:oracle.run1.framed-oracle.run2.framed\",\n \"device\": \"cuda\",\n \"lr\": 0.0001,\n \"n_shot\": 3,\n \"n_query\": 2,\n \"train_k_factor\": 3,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_accuracy\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"n_way\": 16,\n \"datasets\": [\n {\n \"labels\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"domains\": [32, 38, 8, 44, 14, 50, 20, 26],\n \"num_examples_per_domain_per_label\": 2000,\n \"pickle_path\": \"/root/csc500-main/datasets/oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_power\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE.run1_\",\n },\n {\n \"labels\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"domains\": [32, 38, 8, 44, 14, 50, 20, 26],\n \"num_examples_per_domain_per_label\": 2000,\n \"pickle_path\": \"/root/csc500-main/datasets/oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_power\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE.run2_\",\n },\n ],\n \"dataset_seed\": 500,\n \"seed\": 500,\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nif \"x_shape\" not in p:\n p.x_shape = [2,256] # Default to this if we dont supply x_shape\n\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"p.domains_source = []\np.domains_target = []\n\n\ntrain_original_source = []\nval_original_source = []\ntest_original_source = []\n\ntrain_original_target = []\nval_original_target = []\ntest_original_target = []",
"_____no_output_____"
],
[
"# global_x_transform_func = lambda x: normalize(x.to(torch.get_default_dtype()), \"unit_power\") # unit_power, unit_mag\n# global_x_transform_func = lambda x: normalize(x, \"unit_power\") # unit_power, unit_mag",
"_____no_output_____"
],
[
"def add_dataset(\n labels,\n domains,\n pickle_path,\n x_transforms,\n episode_transforms,\n domain_prefix,\n num_examples_per_domain_per_label,\n source_or_target_dataset:str,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n):\n \n if x_transforms == []: x_transform = None\n else: x_transform = get_chained_transform(x_transforms)\n \n if episode_transforms == []: episode_transform = None\n else: raise Exception(\"episode_transforms not implemented\")\n \n episode_transform = lambda tup, _prefix=domain_prefix: (_prefix + str(tup[0]), tup[1])\n\n\n eaf = Episodic_Accessor_Factory(\n labels=labels,\n domains=domains,\n num_examples_per_domain_per_label=num_examples_per_domain_per_label,\n iterator_seed=iterator_seed,\n dataset_seed=dataset_seed,\n n_shot=n_shot,\n n_way=n_way,\n n_query=n_query,\n train_val_test_k_factors=train_val_test_k_factors,\n pickle_path=pickle_path,\n x_transform_func=x_transform,\n )\n\n train, val, test = eaf.get_train(), eaf.get_val(), eaf.get_test()\n train = Lazy_Iterable_Wrapper(train, episode_transform)\n val = Lazy_Iterable_Wrapper(val, episode_transform)\n test = Lazy_Iterable_Wrapper(test, episode_transform)\n\n if source_or_target_dataset==\"source\":\n train_original_source.append(train)\n val_original_source.append(val)\n test_original_source.append(test)\n\n p.domains_source.extend(\n [domain_prefix + str(u) for u in domains]\n )\n elif source_or_target_dataset==\"target\":\n train_original_target.append(train)\n val_original_target.append(val)\n test_original_target.append(test)\n p.domains_target.extend(\n [domain_prefix + str(u) for u in domains]\n )\n else:\n raise Exception(f\"invalid source_or_target_dataset: {source_or_target_dataset}\")\n ",
"_____no_output_____"
],
[
"for ds in p.datasets:\n add_dataset(**ds)",
"_____no_output_____"
],
[
"# from steves_utils.CORES.utils import (\n# ALL_NODES,\n# ALL_NODES_MINIMUM_1000_EXAMPLES,\n# ALL_DAYS\n# )\n\n# add_dataset(\n# labels=ALL_NODES,\n# domains = ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"cores_{u}\"\n# )",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle1_{u}\"\n# )\n",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62,56}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle2_{u}\"\n# )",
"_____no_output_____"
],
[
"# add_dataset(\n# labels=list(range(19)),\n# domains = [0,1,2],\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"metehan.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"met_{u}\"\n# )",
"_____no_output_____"
],
[
"# # from steves_utils.wisig.utils import (\n# # ALL_NODES_MINIMUM_100_EXAMPLES,\n# # ALL_NODES_MINIMUM_500_EXAMPLES,\n# # ALL_NODES_MINIMUM_1000_EXAMPLES,\n# # ALL_DAYS\n# # )\n\n# import steves_utils.wisig.utils as wisig\n\n\n# add_dataset(\n# labels=wisig.ALL_NODES_MINIMUM_100_EXAMPLES,\n# domains = wisig.ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"wisig.node3-19.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"wisig_{u}\"\n# )",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\ntrain_original_source = Iterable_Aggregator(train_original_source, p.seed)\nval_original_source = Iterable_Aggregator(val_original_source, p.seed)\ntest_original_source = Iterable_Aggregator(test_original_source, p.seed)\n\n\ntrain_original_target = Iterable_Aggregator(train_original_target, p.seed)\nval_original_target = Iterable_Aggregator(val_original_target, p.seed)\ntest_original_target = Iterable_Aggregator(test_original_target, p.seed)\n\n# For CNN We only use X and Y. And we only train on the source.\n# Properly form the data using a transform lambda and Lazy_Iterable_Wrapper. Finally wrap them in a dataloader\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"from steves_utils.transforms import get_average_magnitude, get_average_power\n\nprint(set([u for u,_ in val_original_source]))\nprint(set([u for u,_ in val_original_target]))\n\ns_x, s_y, q_x, q_y, _ = next(iter(train_processed_source))\nprint(s_x)\n\n# for ds in [\n# train_processed_source,\n# val_processed_source,\n# test_processed_source,\n# train_processed_target,\n# val_processed_target,\n# test_processed_target\n# ]:\n# for s_x, s_y, q_x, q_y, _ in ds:\n# for X in (s_x, q_x):\n# for x in X:\n# assert np.isclose(get_average_magnitude(x.numpy()), 1.0)\n# assert np.isclose(get_average_power(x.numpy()), 1.0)\n ",
"{'ORACLE.run2_14', 'ORACLE.run2_32', 'ORACLE.run2_26', 'ORACLE.run2_44', 'ORACLE.run2_50', 'ORACLE.run2_38', 'ORACLE.run2_20', 'ORACLE.run2_8'}\n"
],
[
"###################################\n# Build the model\n###################################\n# easfsl only wants a tuple for the shape\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=tuple(p.x_shape))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 256)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 6720], examples_per_second: 33.5196, train_label_loss: 2.8330, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.8950846354166667 Target Test Label Accuracy: 0.8846354166666667\nSource Val Label Accuracy: 0.8943684895833334 Target Val Label Accuracy: 0.8854166666666666\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b7660a9601297d31ef9744f123e404e10754d0 | 1,803 | ipynb | Jupyter Notebook | docs/contents/tools/files/file_pdb/to_pytraj_Topology.ipynb | dprada/molsysmt | 83f150bfe3cfa7603566a0ed4aed79d9b0c97f5d | [
"MIT"
] | null | null | null | docs/contents/tools/files/file_pdb/to_pytraj_Topology.ipynb | dprada/molsysmt | 83f150bfe3cfa7603566a0ed4aed79d9b0c97f5d | [
"MIT"
] | null | null | null | docs/contents/tools/files/file_pdb/to_pytraj_Topology.ipynb | dprada/molsysmt | 83f150bfe3cfa7603566a0ed4aed79d9b0c97f5d | [
"MIT"
] | null | null | null | 20.033333 | 84 | 0.537992 | [
[
[
"# To pytraj.Topology",
"_____no_output_____"
]
],
[
[
"from molsysmt.tools import file_pdb",
"Warning: importing 'simtk.openmm' is deprecated. Import 'openmm' instead.\n"
],
[
"#file_pdb.to_pytraj_Topology(item)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
]
] |
d0b76afe219eb01c37896225e24f11c064de7405 | 9,434 | ipynb | Jupyter Notebook | docs/source/demos/fraud.ipynb | freddyaboulton/evalml | 53242f9d7397a1af3f8c48d498a023042790d4c3 | [
"BSD-3-Clause"
] | 454 | 2020-09-25T15:36:06.000Z | 2022-03-30T04:48:49.000Z | docs/source/demos/fraud.ipynb | freddyaboulton/evalml | 53242f9d7397a1af3f8c48d498a023042790d4c3 | [
"BSD-3-Clause"
] | 2,175 | 2020-09-25T17:05:45.000Z | 2022-03-31T19:54:54.000Z | docs/source/demos/fraud.ipynb | isabella232/evalml | 5b372d0dfac05ff9b7e41eb494a9df1bf2da4a9d | [
"BSD-3-Clause"
] | 66 | 2020-09-25T18:46:27.000Z | 2022-03-02T18:33:30.000Z | 31.032895 | 457 | 0.596036 | [
[
[
"# Building a Fraud Prediction Model with EvalML\n\nIn this demo, we will build an optimized fraud prediction model using EvalML. To optimize the pipeline, we will set up an objective function to minimize the percentage of total transaction value lost to fraud. At the end of this demo, we also show you how introducing the right objective during the training results in a much better than using a generic machine learning metric like AUC.",
"_____no_output_____"
]
],
[
[
"import evalml\nfrom evalml import AutoMLSearch\nfrom evalml.objectives import FraudCost",
"_____no_output_____"
]
],
[
[
"## Configure \"Cost of Fraud\" \n\nTo optimize the pipelines toward the specific business needs of this model, we can set our own assumptions for the cost of fraud. These parameters are\n\n* `retry_percentage` - what percentage of customers will retry a transaction if it is declined?\n* `interchange_fee` - how much of each successful transaction do you collect?\n* `fraud_payout_percentage` - the percentage of fraud will you be unable to collect\n* `amount_col` - the column in the data the represents the transaction amount\n\nUsing these parameters, EvalML determines attempt to build a pipeline that will minimize the financial loss due to fraud.",
"_____no_output_____"
]
],
[
[
"fraud_objective = FraudCost(retry_percentage=.5,\n interchange_fee=.02,\n fraud_payout_percentage=.75,\n amount_col='amount')",
"_____no_output_____"
]
],
[
[
"## Search for best pipeline",
"_____no_output_____"
],
[
"In order to validate the results of the pipeline creation and optimization process, we will save some of our data as the holdout set.",
"_____no_output_____"
]
],
[
[
"X, y = evalml.demos.load_fraud(n_rows=5000)",
"_____no_output_____"
]
],
[
[
"EvalML natively supports one-hot encoding. Here we keep 1 out of the 6 categorical columns to decrease computation time.",
"_____no_output_____"
]
],
[
[
"cols_to_drop = ['datetime', 'expiration_date', 'country', 'region', 'provider']\nfor col in cols_to_drop:\n X.ww.pop(col)\n\nX_train, X_holdout, y_train, y_holdout = evalml.preprocessing.split_data(X, y, problem_type='binary', test_size=0.2, random_seed=0)\n\nX.ww",
"_____no_output_____"
]
],
[
[
"Because the fraud labels are binary, we will use `AutoMLSearch(X_train=X_train, y_train=y_train, problem_type='binary')`. When we call `.search()`, the search for the best pipeline will begin. ",
"_____no_output_____"
]
],
[
[
"automl = AutoMLSearch(X_train=X_train, y_train=y_train,\n problem_type='binary', \n objective=fraud_objective,\n additional_objectives=['auc', 'f1', 'precision'],\n allowed_model_families=[\"random_forest\", \"linear_model\"],\n max_batches=1,\n optimize_thresholds=True,\n verbose=True)\n\nautoml.search()",
"_____no_output_____"
]
],
[
[
"### View rankings and select pipelines\n\nOnce the fitting process is done, we can see all of the pipelines that were searched, ranked by their score on the fraud detection objective we defined.",
"_____no_output_____"
]
],
[
[
"automl.rankings",
"_____no_output_____"
]
],
[
[
"To select the best pipeline we can call `automl.best_pipeline`.",
"_____no_output_____"
]
],
[
[
"best_pipeline = automl.best_pipeline",
"_____no_output_____"
]
],
[
[
"### Describe pipelines\n\nWe can get more details about any pipeline created during the search process, including how it performed on other objective functions, by calling the `describe_pipeline` method and passing the `id` of the pipeline of interest.",
"_____no_output_____"
]
],
[
[
"automl.describe_pipeline(automl.rankings.iloc[1][\"id\"])",
"_____no_output_____"
]
],
[
[
"## Evaluate on holdout data\n\nFinally, since the best pipeline is already trained, we evaluate it on the holdout data.",
"_____no_output_____"
],
[
"Now, we can score the pipeline on the holdout data using both our fraud cost objective and the AUC (Area under the ROC Curve) objective.",
"_____no_output_____"
]
],
[
[
"best_pipeline.score(X_holdout, y_holdout, objectives=[\"auc\", fraud_objective])",
"_____no_output_____"
]
],
[
[
"## Why optimize for a problem-specific objective?\n\nTo demonstrate the importance of optimizing for the right objective, let's search for another pipeline using AUC, a common machine learning metric. After that, we will score the holdout data using the fraud cost objective to see how the best pipelines compare.",
"_____no_output_____"
]
],
[
[
"automl_auc = AutoMLSearch(X_train=X_train, y_train=y_train,\n problem_type='binary',\n objective='auc',\n additional_objectives=['f1', 'precision'],\n max_batches=1,\n allowed_model_families=[\"random_forest\", \"linear_model\"],\n optimize_thresholds=True,\n verbose=True)\n\nautoml_auc.search()",
"_____no_output_____"
]
],
[
[
"Like before, we can look at the rankings of all of the pipelines searched and pick the best pipeline.",
"_____no_output_____"
]
],
[
[
"automl_auc.rankings",
"_____no_output_____"
],
[
"best_pipeline_auc = automl_auc.best_pipeline",
"_____no_output_____"
],
[
"# get the fraud score on holdout data\nbest_pipeline_auc.score(X_holdout, y_holdout, objectives=[\"auc\", fraud_objective])",
"_____no_output_____"
],
[
"# fraud score on fraud optimized again\nbest_pipeline.score(X_holdout, y_holdout, objectives=[\"auc\", fraud_objective])",
"_____no_output_____"
]
],
[
[
"When we optimize for AUC, we can see that the AUC score from this pipeline performs better compared to the AUC score from the pipeline optimized for fraud cost; however, the losses due to fraud are a much larger percentage of the total transaction amount when optimized for AUC and much smaller when optimized for fraud cost. As a result, we lose a noticable percentage of the total transaction amount by not optimizing for fraud cost specifically.\n\nOptimizing for AUC does not take into account the user-specified `retry_percentage`, `interchange_fee`, `fraud_payout_percentage` values, which could explain the decrease in fraud performance. Thus, the best pipelines may produce the highest AUC but may not actually reduce the amount loss due to your specific type fraud.\n\nThis example highlights how performance in the real world can diverge greatly from machine learning metrics.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0b7761f53929cfef67225c028a9e043c6902c95 | 10,817 | ipynb | Jupyter Notebook | notebooks/cvr/Medarbdstal.ipynb | mssalvador/notebooks | 7b6d0619f4e62cec7ef953ed9330d4a1ee15e7c8 | [
"Apache-2.0"
] | null | null | null | notebooks/cvr/Medarbdstal.ipynb | mssalvador/notebooks | 7b6d0619f4e62cec7ef953ed9330d4a1ee15e7c8 | [
"Apache-2.0"
] | null | null | null | notebooks/cvr/Medarbdstal.ipynb | mssalvador/notebooks | 7b6d0619f4e62cec7ef953ed9330d4a1ee15e7c8 | [
"Apache-2.0"
] | null | null | null | 37.689895 | 169 | 0.552279 | [
[
[
"#Always Pyspark first!\nErhvervsPath = \"/home/svanhmic/workspace/Python/Erhvervs\"\n\nfrom pyspark.sql import functions as F, Window, WindowSpec\nfrom pyspark.sql import Row\nfrom pyspark.sql.types import StringType,ArrayType,IntegerType,DoubleType,StructField,StructType\nsc.addPyFile(ErhvervsPath+\"/src/RegnSkabData/ImportRegnskabData.py\")\nsc.addPyFile(ErhvervsPath+'/src/RegnSkabData/RegnskabsClass.py')\nsc.addPyFile(ErhvervsPath+'/src/cvr/GetNextJsonLayer.py')\n\nimport sys\nimport re\nimport os\nimport ImportRegnskabData\nimport GetNextJsonLayer\nimport itertools\nimport functools\n\n",
"_____no_output_____"
],
[
"cvrPath = \"/home/svanhmic/workspace/Python/Erhvervs/data/cdata/parquet\"\ncvrfiles = os.listdir(cvrPath)",
"_____no_output_____"
],
[
"#import crv data\ncvrDf = (sqlContext\n .read\n .parquet(cvrPath+\"/\"+cvrfiles[0])\n )\n#cvrDf.show(1)\n#print(cvrDf.select(\"cvrNummer\").distinct().count())\n\n#Extract all Aps and A/S companies\nvirkformCols = (\"cvrNummer\",\"virksomhedsform\")\n\nvirkformDf = GetNextJsonLayer.createNextLayerTable(cvrDf.select(*virkformCols),[virkformCols[0]],virkformCols[1])\nvirkformDf = GetNextJsonLayer.expandSubCols(virkformDf,mainColumn=\"periode\")\nvirkformDf = (virkformDf\n .drop(\"sidstOpdateret\")\n .withColumn(col=F.col(\"periode_gyldigFra\").cast(\"date\"),colName=\"periode_gyldigFra\")\n .withColumn(col=F.col(\"periode_gyldigTil\").cast(\"date\"),colName=\"periode_gyldigTil\")\n )\n\n#virkformDf.show(1)\ncheckCols = [\"kortBeskrivelse\",\"langBeskrivelse\",\"virksomhedsformkode\"]\n\n#Consistencycheck is kortBeskrivelse and virksomhedsformkode always mapped the same way\n#check1 = virkformDf.select(checkCols+[\"cvrNummer\"]).distinct().groupby(*checkCols).count()\n#check1.orderBy(\"kortBeskrivelse\",\"count\").show(check1.count(),truncate=False)\n\n#Second test does any companies go from Aps or A/S to other or vice versa?\njoinCols = [\"cvrNummer\",\"langBeskrivelse\",\"rank\"]\ncvrCols = [\"cvrNummer\"]\ngyldigCol = [\"periode_gyldigFra\"]\n\nstatusChangeWindow = (Window\n .partitionBy(F.col(*cvrCols))\n .orderBy(F.col(\"periode_gyldigFra\").desc()))\n\n#virkformDf.select(checkCols).distinct().show(50,truncate=False)\n\n\n\n#Extract APS and AS here and latest status...\naggregationCols = [F.max(i) for i in gyldigCol]\ngroupsCol = [i for i in virkformDf.columns if i not in gyldigCol]\n\ncompanyByAsApsDf = (virkformDf\n .where((F.col(\"virksomhedsformkode\") == 60) | (F.col(\"virksomhedsformkode\") == 80))\n .withColumn(col=F.rank().over(statusChangeWindow),colName=\"rank\")\n .filter(F.col(\"rank\") == 1)\n )\n",
"_____no_output_____"
],
[
"#Get the medarbejdstal\nfastCols = [\"cvrNummer\",\"aar\"]\nregCols = [\"intervalKodeAntalAarsvaerk\",\"intervalKodeAntalInklusivEjere\"]\nreg2Cols = [\"intervalKodeAntalAarsvaerk\",\"intervalKodeAntalAnsatte\"]\n\nfCols = [F.split(F.regexp_replace(F.col(i),pattern=r'ANTAL_',replacement=\"\"),\"_\").alias(i) for i in regCols]\nmkCols = [F.split(F.regexp_replace(F.col(i),pattern=r'ANTAL_',replacement=\"\"),\"_\").alias(i) for i in reg2Cols]\n#kvartCols = [F.split(F.regexp_replace(F.col(i),pattern=r'ANTAL_',replacement=\"\"),\"_\").alias(i) for i in cols]\n\ndef getLower(x):\n \n try:\n return int(x[0])\n except:\n return None\n\ndef getUpper(x):\n \n try:\n return int(x[0])\n except:\n return None\n\ngetLowerBound = F.udf(lambda x: getLower(x),IntegerType())\ngetUpperBound = F.udf(lambda x: getUpper(x),IntegerType())\n\naarsDf = (GetNextJsonLayer\n .createNextLayerTable(cvrDf,[\"cvrNummer\"],\"aarsbeskaeftigelse\")\n .select(fastCols+fCols)\n .select(fastCols+[getLowerBound(i).alias(\"lower_\"+i) for i in regCols]) \n )\n\nmaanedsDf = (GetNextJsonLayer\n .createNextLayerTable(cvrDf,[\"cvrNummer\"],\"maanedsbeskaeftigelse\")\n .select([\"cvrNummer\",\"aar\",\"maaned\"]+mkCols)\n .select([\"cvrNummer\",\"aar\",\"maaned\"]+[getLowerBound(i).alias(\"lower_\"+i) for i in reg2Cols])\n )\n \nkvartDf = (GetNextJsonLayer\n .createNextLayerTable(cvrDf,[\"cvrNummer\"],\"kvartalsbeskaeftigelse\")\n .select([\"cvrNummer\",\"aar\",\"kvartal\"]+mkCols)\n .select([\"cvrNummer\",\"aar\",\"kvartal\"]+[getLowerBound(i).alias(\"lower_\"+i) for i in reg2Cols])\n )\n\n#maanedsDf.show()\n#cvrDf.unpersist()\n#maanedsDf.show()\n#kvartDf.show()\n#print(aarsDf.count())\n#print(aarsDf.na.drop(how=\"all\",subset=[\"lower_\"+i for i in cols]).count())",
"_____no_output_____"
],
[
"# OK how many are represented in both or all three groups? \ndistinctMaanedDf = (maanedsDf\n .join(companyByAsApsDf,on=(maanedsDf[\"cvrNummer\"]==companyByAsApsDf[\"cvrNummer\"]),how=\"right\")\n .drop(companyByAsApsDf[\"cvrNummer\"])\n .distinct()\n \n )\n#distinctMaanedDf.show()\n\ndistinctKvartalDf = (kvartDf \n .join(companyByAsApsDf,on=(kvartDf[\"cvrNummer\"]==companyByAsApsDf[\"cvrNummer\"]),how=\"right\")\n .drop(companyByAsApsDf[\"cvrNummer\"])\n .distinct()\n )\n\ndistinctAarDf = (aarsDf\n .join(companyByAsApsDf,on=(aarsDf[\"cvrNummer\"]==companyByAsApsDf[\"cvrNummer\"]),how=\"right\")\n .drop(companyByAsApsDf[\"cvrNummer\"])\n .distinct()\n )\n\n",
"_____no_output_____"
],
[
"distinctMaanedDf.write.parquet(mode=\"overwrite\",path=cvrPath+\"/MaanedsVaerker.parquet\")\ndistinctKvartalDf.write.parquet(mode=\"overwrite\",path=cvrPath+\"/KvartalsVaerker.parquet\")\ndistinctAarDf.write.parquet(mode=\"overwrite\",path=cvrPath+\"/AarsVaerker.parquet\")\n\n#print(\"månedsbeskæftigelse: \"+str(distinctMaanedDf.count()))\n#print(\"kvartalsbeskæftigelse: \"+str(distinctKvartalDf.count()))\n#print(\"årsbeskæftigelse: \"+str(distinctAarDf.count()))\n\n\n\n#print(\"Årsbeskæftigelse til kvartalsbeskæftigelse: \"+str(distinctAarDf.select(F.col(\"cvrNummer\")).distinct()\n# .join(distinctKvartalDf.select(F.col(\"cvrNummer\")).distinct(),(distinctKvartalDf[\"cvrNummer\"]==distinctAarDf[\"cvrNummer\"]),how=\"inner\")\n# .drop(distinctAarDf[\"cvrNummer\"]).distinct().count()\n#))\n#print(\"Årsbeskæftigelse til månedsbeskæftigelse: \"+str(distinctAarDf.select(F.col(\"cvrNummer\")).distinct()\n# .join(distinctMaanedDf.select(F.col(\"cvrNummer\")).distinct(),(distinctMaanedDf[\"cvrNummer\"]==distinctAarDf[\"cvrNummer\"]),how=\"inner\")\n# .drop(distinctAarDf[\"cvrNummer\"]).distinct().count()\n#))\n#print(\"Kvartalsbeskæftigelse til månedsbeskæftigelse: \"+str(distinctKvartalDf.select(F.col(\"cvrNummer\")).distinct()\n# .join(distinctMaanedDf.select(F.col(\"cvrNummer\")).distinct(),(distinctMaanedDf[\"cvrNummer\"]==distinctKvartalDf[\"cvrNummer\"]),how=\"inner\")\n# .drop(distinctAarDf[\"cvrNummer\"]).distinct().count()\n#))\n\nAllThreeCount = (distinctAarDf\n .select(F.col(\"cvrNummer\")).distinct()\n .join(distinctKvartalDf.select(F.col(\"cvrNummer\")).distinct(),(distinctAarDf[\"cvrNummer\"]==distinctKvartalDf[\"cvrNummer\"]),how=\"inner\")\n .drop(distinctKvartalDf[\"cvrNummer\"])\n .join(distinctMaanedDf.select(F.col(\"cvrNummer\")).distinct(),(distinctAarDf[\"cvrNummer\"]==distinctMaanedDf[\"cvrNummer\"]),how=\"inner\")\n .drop(distinctMaanedDf[\"cvrNummer\"])\n .distinct()\n )\n#print(\"Aarsbeskæftigelse til kvartalsbeskæftigelse til månedsbeskæftigelse: \"+str(AllThreeCount.count()))",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b789f075ff9da2c79f1dce92b12e6317ac81ba | 18,284 | ipynb | Jupyter Notebook | ModeloDeAhorro.ipynb | Rubn01/simlacion | 2974218bc9cc8ba91e726fe0b05eac76a9e6c608 | [
"MIT"
] | null | null | null | ModeloDeAhorro.ipynb | Rubn01/simlacion | 2974218bc9cc8ba91e726fe0b05eac76a9e6c608 | [
"MIT"
] | null | null | null | ModeloDeAhorro.ipynb | Rubn01/simlacion | 2974218bc9cc8ba91e726fe0b05eac76a9e6c608 | [
"MIT"
] | null | null | null | 78.136752 | 6,068 | 0.810982 | [
[
[
"### Ruben Arias",
"_____no_output_____"
],
[
">\n- Simple \n- Compuesto \n- Continuo",
"_____no_output_____"
]
],
[
[
"# interes, i, C0, capital inicial, r, periodos\ndef intSimpleCapital(capital, periodos, interes):\n return capital * (1 + periodos * interes)",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt \n%matplotlib inline",
"_____no_output_____"
],
[
"listaDeInteres = [.05,.1, .15, .2, .5]",
"_____no_output_____"
],
[
"# capital = 10000\nperiodos = 12\nfor interes in listaDeInteres:\n print(intSimpleCapital(10000, periodos, interes), end = \" \")",
"16000.0 22000.0 28000.0 34000.0 70000.0 "
],
[
"periodos = [3, 6, 12, 18]\ncolores = [\"black\", \"red\", \"blue\", \"orange\"]\nplt.figure(figsize = (4,2))\nfor indx, periodo in enumerate(periodos):\n for interes in listaDeInteres:\n capitalx = intSimpleCapital(10000, periodo, interes)\n plt.scatter([periodo], [capitalx], color = colores[indx], s = 5)",
"_____no_output_____"
],
[
"def intCompuestoCapital(capital, periodos, interes):\n return capital * (1 + interes)**periodos",
"_____no_output_____"
],
[
"periodos = 12\nfor interes in listaDeInteres:\n print(intCompuestoCapital(10000, periodos, interes), end = \" \")",
"_____no_output_____"
],
[
"periodos = [3, 6, 12, 18]\ncolores = [\"black\", \"red\", \"blue\", \"orange\"]\nplt.figure(figsize = (4,2))\nfor indx, periodo in enumerate(periodos):\n for interes in listaDeInteres:\n capitalx = intSimpleCapital(1000, periodo, interes)\n plt.scatter([periodo], [capitalx], color = colores[indx], s = 5)",
"_____no_output_____"
],
[
"def intContinuo():\n return",
"_____no_output_____"
],
[
"def intSimpleCapital(capital, periodos, interes):\n return capital * (1 + periodos * interes)",
"_____no_output_____"
],
[
"C = c0 (1 - i)**periodos",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b78d46b894399e2d028f497cfcb9207ebde1ff | 21,784 | ipynb | Jupyter Notebook | site/en/guide/keras/custom_callback.ipynb | bogdan-marian/tensorflow-docs | 10f4ff366fe532cd397eb5f44537e74dcc666d10 | [
"Apache-2.0"
] | 3 | 2020-01-28T11:36:06.000Z | 2020-01-28T12:15:04.000Z | site/en/guide/keras/custom_callback.ipynb | bogdan-marian/tensorflow-docs | 10f4ff366fe532cd397eb5f44537e74dcc666d10 | [
"Apache-2.0"
] | 1 | 2020-02-20T14:49:33.000Z | 2020-02-20T14:49:33.000Z | site/en/guide/keras/custom_callback.ipynb | bogdan-marian/tensorflow-docs | 10f4ff366fe532cd397eb5f44537e74dcc666d10 | [
"Apache-2.0"
] | 1 | 2020-03-04T00:12:25.000Z | 2020-03-04T00:12:25.000Z | 38.83066 | 695 | 0.545813 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/keras/custom_callback\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/keras/custom_callback.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/guide/keras/custom_callback.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/keras/custom_callback.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"# Keras custom callbacks\nA custom callback is a powerful tool to customize the behavior of a Keras model during training, evaluation, or inference, including reading/changing the Keras model. Examples include `tf.keras.callbacks.TensorBoard` where the training progress and results can be exported and visualized with TensorBoard, or `tf.keras.callbacks.ModelCheckpoint` where the model is automatically saved during training, and more. In this guide, you will learn what Keras callback is, when it will be called, what it can do, and how you can build your own. Towards the end of this guide, there will be demos of creating a couple of simple callback applications to get you started on your custom callback.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\n\ntry:\n # %tensorflow_version only exists in Colab.\n %tensorflow_version 2.x\nexcept Exception:\n pass\nimport tensorflow as tf",
"_____no_output_____"
]
],
[
[
"## Introduction to Keras callbacks\nIn Keras, `Callback` is a python class meant to be subclassed to provide specific functionality, with a set of methods called at various stages of training (including batch/epoch start and ends), testing, and predicting. Callbacks are useful to get a view on internal states and statistics of the model during training. You can pass a list of callbacks (as the keyword argument `callbacks`) to any of `tf.keras.Model.fit()`, `tf.keras.Model.evaluate()`, and `tf.keras.Model.predict()` methods. The methods of the callbacks will then be called at different stages of training/evaluating/inference.\n\nTo get started, let's import tensorflow and define a simple Sequential Keras model:",
"_____no_output_____"
]
],
[
[
"# Define the Keras model to add callbacks to\ndef get_model():\n model = tf.keras.Sequential()\n model.add(tf.keras.layers.Dense(1, activation = 'linear', input_dim = 784))\n model.compile(optimizer=tf.keras.optimizers.RMSprop(lr=0.1), loss='mean_squared_error', metrics=['mae'])\n return model",
"_____no_output_____"
]
],
[
[
"Then, load the MNIST data for training and testing from Keras datasets API:",
"_____no_output_____"
]
],
[
[
"# Load example MNIST data and pre-process it\n(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()\nx_train = x_train.reshape(60000, 784).astype('float32') / 255\nx_test = x_test.reshape(10000, 784).astype('float32') / 255",
"_____no_output_____"
]
],
[
[
"Now, define a simple custom callback to track the start and end of every batch of data. During those calls, it prints the index of the current batch.",
"_____no_output_____"
]
],
[
[
"import datetime\n\nclass MyCustomCallback(tf.keras.callbacks.Callback):\n\n def on_train_batch_begin(self, batch, logs=None):\n print('Training: batch {} begins at {}'.format(batch, datetime.datetime.now().time()))\n\n def on_train_batch_end(self, batch, logs=None):\n print('Training: batch {} ends at {}'.format(batch, datetime.datetime.now().time()))\n\n def on_test_batch_begin(self, batch, logs=None):\n print('Evaluating: batch {} begins at {}'.format(batch, datetime.datetime.now().time()))\n\n def on_test_batch_end(self, batch, logs=None):\n print('Evaluating: batch {} ends at {}'.format(batch, datetime.datetime.now().time()))",
"_____no_output_____"
]
],
[
[
"Providing a callback to model methods such as `tf.keras.Model.fit()` ensures the methods are called at those stages:",
"_____no_output_____"
]
],
[
[
"model = get_model()\n_ = model.fit(x_train, y_train,\n batch_size=64,\n epochs=1,\n steps_per_epoch=5,\n verbose=0,\n callbacks=[MyCustomCallback()])",
"_____no_output_____"
]
],
[
[
"## Model methods that take callbacks\nUsers can supply a list of callbacks to the following `tf.keras.Model` methods:\n#### [`fit()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit), [`fit_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#fit_generator)\nTrains the model for a fixed number of epochs (iterations over a dataset, or data yielded batch-by-batch by a Python generator).\n#### [`evaluate()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#evaluate), [`evaluate_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#evaluate_generator)\nEvaluates the model for given data or data generator. Outputs the loss and metric values from the evaluation.\n#### [`predict()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#predict), [`predict_generator()`](https://www.tensorflow.org/api_docs/python/tf/keras/Model#predict_generator)\nGenerates output predictions for the input data or data generator.\n",
"_____no_output_____"
]
],
[
[
"_ = model.evaluate(x_test, y_test, batch_size=128, verbose=0, steps=5,\n callbacks=[MyCustomCallback()])",
"_____no_output_____"
]
],
[
[
"## An overview of callback methods\n\n\n### Common methods for training/testing/predicting\nFor training, testing, and predicting, following methods are provided to be overridden.\n#### `on_(train|test|predict)_begin(self, logs=None)`\nCalled at the beginning of `fit`/`evaluate`/`predict`.\n#### `on_(train|test|predict)_end(self, logs=None)`\nCalled at the end of `fit`/`evaluate`/`predict`.\n#### `on_(train|test|predict)_batch_begin(self, batch, logs=None)`\nCalled right before processing a batch during training/testing/predicting. Within this method, `logs` is a dict with `batch` and `size` available keys, representing the current batch number and the size of the batch.\n#### `on_(train|test|predict)_batch_end(self, batch, logs=None)`\nCalled at the end of training/testing/predicting a batch. Within this method, `logs` is a dict containing the stateful metrics result.\n\n### Training specific methods\nIn addition, for training, following are provided.\n#### on_epoch_begin(self, epoch, logs=None)\nCalled at the beginning of an epoch during training.\n#### on_epoch_end(self, epoch, logs=None)\nCalled at the end of an epoch during training.\n",
"_____no_output_____"
],
[
"### Usage of `logs` dict\nThe `logs` dict contains the loss value, and all the metrics at the end of a batch or epoch. Example includes the loss and mean absolute error.",
"_____no_output_____"
]
],
[
[
"class LossAndErrorPrintingCallback(tf.keras.callbacks.Callback):\n\n def on_train_batch_end(self, batch, logs=None):\n print('For batch {}, loss is {:7.2f}.'.format(batch, logs['loss']))\n\n def on_test_batch_end(self, batch, logs=None):\n print('For batch {}, loss is {:7.2f}.'.format(batch, logs['loss']))\n\n def on_epoch_end(self, epoch, logs=None):\n print('The average loss for epoch {} is {:7.2f} and mean absolute error is {:7.2f}.'.format(epoch, logs['loss'], logs['mae']))\n\nmodel = get_model()\n_ = model.fit(x_train, y_train,\n batch_size=64,\n steps_per_epoch=5,\n epochs=3,\n verbose=0,\n callbacks=[LossAndErrorPrintingCallback()])",
"_____no_output_____"
]
],
[
[
"Similarly, one can provide callbacks in `evaluate()` calls.",
"_____no_output_____"
]
],
[
[
"_ = model.evaluate(x_test, y_test, batch_size=128, verbose=0, steps=20,\n callbacks=[LossAndErrorPrintingCallback()])",
"_____no_output_____"
]
],
[
[
"## Examples of Keras callback applications\nThe following section will guide you through creating simple Callback applications.",
"_____no_output_____"
],
[
"### Early stopping at minimum loss\nFirst example showcases the creation of a `Callback` that stops the Keras training when the minimum of loss has been reached by mutating the attribute `model.stop_training` (boolean). Optionally, the user can provide an argument `patience` to specify how many epochs the training should wait before it eventually stops.\n\n`tf.keras.callbacks.EarlyStopping` provides a more complete and general implementation.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nclass EarlyStoppingAtMinLoss(tf.keras.callbacks.Callback):\n \"\"\"Stop training when the loss is at its min, i.e. the loss stops decreasing.\n\n Arguments:\n patience: Number of epochs to wait after min has been hit. After this\n number of no improvement, training stops.\n \"\"\"\n\n def __init__(self, patience=0):\n super(EarlyStoppingAtMinLoss, self).__init__()\n\n self.patience = patience\n\n # best_weights to store the weights at which the minimum loss occurs.\n self.best_weights = None\n\n def on_train_begin(self, logs=None):\n # The number of epoch it has waited when loss is no longer minimum.\n self.wait = 0\n # The epoch the training stops at.\n self.stopped_epoch = 0\n # Initialize the best as infinity.\n self.best = np.Inf\n\n def on_epoch_end(self, epoch, logs=None):\n current = logs.get('loss')\n if np.less(current, self.best):\n self.best = current\n self.wait = 0\n # Record the best weights if current results is better (less).\n self.best_weights = self.model.get_weights()\n else:\n self.wait += 1\n if self.wait >= self.patience:\n self.stopped_epoch = epoch\n self.model.stop_training = True\n print('Restoring model weights from the end of the best epoch.')\n self.model.set_weights(self.best_weights)\n\n def on_train_end(self, logs=None):\n if self.stopped_epoch > 0:\n print('Epoch %05d: early stopping' % (self.stopped_epoch + 1))",
"_____no_output_____"
],
[
"model = get_model()\n_ = model.fit(x_train, y_train,\n batch_size=64,\n steps_per_epoch=5,\n epochs=30,\n verbose=0,\n callbacks=[LossAndErrorPrintingCallback(), EarlyStoppingAtMinLoss()])",
"_____no_output_____"
]
],
[
[
"### Learning rate scheduling\n\nOne thing that is commonly done in model training is changing the learning rate as more epochs have passed. Keras backend exposes get_value API which can be used to set the variables. In this example, we're showing how a custom Callback can be used to dynamically change the learning rate.\n\nNote: this is just an example implementation see `callbacks.LearningRateScheduler` and `keras.optimizers.schedules` for more general implementations.",
"_____no_output_____"
]
],
[
[
"class LearningRateScheduler(tf.keras.callbacks.Callback):\n \"\"\"Learning rate scheduler which sets the learning rate according to schedule.\n\n Arguments:\n schedule: a function that takes an epoch index\n (integer, indexed from 0) and current learning rate\n as inputs and returns a new learning rate as output (float).\n \"\"\"\n\n def __init__(self, schedule):\n super(LearningRateScheduler, self).__init__()\n self.schedule = schedule\n\n def on_epoch_begin(self, epoch, logs=None):\n if not hasattr(self.model.optimizer, 'lr'):\n raise ValueError('Optimizer must have a \"lr\" attribute.')\n # Get the current learning rate from model's optimizer.\n lr = float(tf.keras.backend.get_value(self.model.optimizer.lr))\n # Call schedule function to get the scheduled learning rate.\n scheduled_lr = self.schedule(epoch, lr)\n # Set the value back to the optimizer before this epoch starts\n tf.keras.backend.set_value(self.model.optimizer.lr, scheduled_lr)\n print('\\nEpoch %05d: Learning rate is %6.4f.' % (epoch, scheduled_lr))",
"_____no_output_____"
],
[
"LR_SCHEDULE = [\n # (epoch to start, learning rate) tuples\n (3, 0.05), (6, 0.01), (9, 0.005), (12, 0.001)\n]\n\ndef lr_schedule(epoch, lr):\n \"\"\"Helper function to retrieve the scheduled learning rate based on epoch.\"\"\"\n if epoch < LR_SCHEDULE[0][0] or epoch > LR_SCHEDULE[-1][0]:\n return lr\n for i in range(len(LR_SCHEDULE)):\n if epoch == LR_SCHEDULE[i][0]:\n return LR_SCHEDULE[i][1]\n return lr\n\nmodel = get_model()\n_ = model.fit(x_train, y_train,\n batch_size=64,\n steps_per_epoch=5,\n epochs=15,\n verbose=0,\n callbacks=[LossAndErrorPrintingCallback(), LearningRateScheduler(lr_schedule)])",
"_____no_output_____"
]
],
[
[
"### Standard Keras callbacks\nBe sure to check out the existing Keras callbacks by [visiting the API doc](https://www.tensorflow.org/api_docs/python/tf/keras/callbacks). Applications include logging to CSV, saving the model, visualizing on TensorBoard and a lot more.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
d0b795c5e23817a77886ddf77675de0461554299 | 3,798 | ipynb | Jupyter Notebook | A_learning_notes/Q&A.ipynb | dywlavender/multi-label-classification | ec8567169b3f1b08c7dafeb71f007275621a5a95 | [
"MIT"
] | 17 | 2019-10-16T04:39:27.000Z | 2022-01-24T03:21:58.000Z | A_learning_notes/Q&A.ipynb | dywlavender/multi-label-classification | ec8567169b3f1b08c7dafeb71f007275621a5a95 | [
"MIT"
] | null | null | null | A_learning_notes/Q&A.ipynb | dywlavender/multi-label-classification | ec8567169b3f1b08c7dafeb71f007275621a5a95 | [
"MIT"
] | 5 | 2020-03-04T09:08:02.000Z | 2021-07-20T02:28:57.000Z | 35.166667 | 333 | 0.541864 | [
[
[
"# Q&A\n- Q. 在编译模型阶段,定义了多输出loss函数和权重,但在训练阶段,打印的loss却不等于各个loss的加权和\n```\nmodel.compile(loss=my_loss, optimizer='adam', loss_weights=[0.5, 0.5])\nmodel.fit(dataset, epochs=2, steps_per_epoch=2, verbose=1)\n```\n输出:loss(21.9610) != 0.5 * 1.3583 - 0.5 * 1.5867\n```\nEpoch 1/2\n1/2 [==============>...............] - ETA: 0s - loss: 21.9610 - dense_4_loss: 1.3583 - dense_5_loss: 1.5867\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\n2/2 [==============================] - 0s 140ms/step - loss: 22.1960 - dense_4_loss: 1.9502 - dense_5_loss: 1.5183\nEpoch 2/2\n1/2 [==============>...............] - ETA: 0s - loss: 21.8526 - dense_4_loss: 1.3555 - dense_5_loss: 1.5861\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\n2/2 [==============================] - 0s 1ms/step - loss: 22.0872 - dense_4_loss: 1.9470 - dense_5_loss: 1.5171\n```\n\n A: 因为总的loss中包含了权重正则化损失部分:\n```\ndef build_net(input_tensor):\n out1 = keras.layers.Dense(1, kernel_initializer='glorot_normal', activation='linear',\n kernel_regularizer=keras.regularizers.l2(10))(input_tensor)\n out2 = keras.layers.Dense(1, kernel_initializer='glorot_normal', activation='linear',\n kernel_regularizer=keras.regularizers.l2(10))(input_tensor)\n return [out1, out2]\n```\n",
"_____no_output_____"
],
[
"- Q. 在将`.ckpt.index` + `.ckpt.data` 模型转为`pb`的时候,为什么还要先保存为`h5`,然后再加载模型,再保存为`pb`?\n\n A: 因为原来保存为`.ckpt.index` + `.ckpt.data` 的时候没有保存图信息,加载也只加载权重信息:\n```\nmodel.load_weights(latest)\n...\ncp_callback = ModelCheckpoint(path, save_weights_only=True, period=ckpt_period)\n```\n导致`keras.backend.get_session().graph.as_graph_def()`没有图结构信息。\n(理论上我是构建了网络图模型,然后再加载权重的,所以应该也得有图结构信息,但实际上没有)\n所以需要将模型完全保存为`h5`(包含图信息),然后重新加载进来,再保存为`pb`:\n```\nmodel.save(h5_path, overwrite=True, include_optimizer=False)\nmodel = keras.models.load_model(h5_path)\n...\ngraph = tf.graph_util.remove_training_nodes(sess.graph.as_graph_def())\ngraph_frozen = tf.graph_util.convert_variables_to_constants(sess, graph, output_names)\ntf.train.write_graph(graph_frozen, pb_model_dir, pb_model_name, as_text=False)\n```",
"_____no_output_____"
],
[
"- Q. 一直没办法用多GPU的模式运行?\n\n A: `tf.enable_eager_execution()`模型跑不了多GPU,要注释掉这句。\n",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
]
] |
d0b7a534ff36949e512dd474134927093899e0ab | 195,022 | ipynb | Jupyter Notebook | python/GAN/GAN.ipynb | alexandre-lavoie/deep-physics | 7727893cef11311e89bf7c030ea09efc25892c6e | [
"MIT"
] | 3 | 2019-03-08T01:04:31.000Z | 2020-09-28T05:45:41.000Z | python/GAN/.ipynb_checkpoints/GAN-checkpoint.ipynb | alexandre-lavoie/deep-physics | 7727893cef11311e89bf7c030ea09efc25892c6e | [
"MIT"
] | null | null | null | python/GAN/.ipynb_checkpoints/GAN-checkpoint.ipynb | alexandre-lavoie/deep-physics | 7727893cef11311e89bf7c030ea09efc25892c6e | [
"MIT"
] | 2 | 2019-03-08T02:03:26.000Z | 2019-12-30T08:12:30.000Z | 213.605696 | 137,116 | 0.871563 | [
[
[
"# Imports",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.models import Model\nfrom tensorflow.keras.layers import Dense, Dropout, Flatten, Input, Concatenate\nfrom tensorflow.keras.optimizers import Adam, RMSprop\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport copy",
"_____no_output_____"
]
],
[
[
"# Global Variables",
"_____no_output_____"
]
],
[
[
"epochs = 500\nbatch_size = 16\nnumber_of_particles = epochs * 2 * batch_size\ndt = 0.1",
"_____no_output_____"
]
],
[
[
"# Classes",
"_____no_output_____"
]
],
[
[
"class Particle:\n def __str__(self):\n return \"Position: %s, Velocity: %s, Accleration: %s\" % (self.position, self.velocity, self.acceleration)\n \n def __repr__(self):\n return \"Position: %s, Velocity: %s, Accleration: %s\" % (self.position, self.velocity, self.acceleration)\n \n def __init__(self):\n self.position = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Position X, Y\n self.velocity = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Velocity X, Y\n self.acceleration = np.array([np.random.sample()*2-1,np.random.sample()*2-1]) # Acceleration X, Y\n \n def apply_physics(self,dt):\n nextParticle = copy.deepcopy(self) # Copy to retain initial values\n nextParticle.position += self.velocity * dt\n nextParticle.velocity += self.acceleration * dt\n return nextParticle\n \n def get_list(self):\n return [self.position[0],self.position[1],self.velocity[0], self.velocity[1], self.acceleration[0], self.acceleration[1]]\n \n def get_list_physics(self,dt):\n n = self.apply_physics(dt)\n return [self.position[0],self.position[1],self.velocity[0], self.velocity[1], self.acceleration[0], \n self.acceleration[1], n.position[0], n.position[1], n.velocity[0], n.velocity[1]]",
"_____no_output_____"
],
[
"class GAN:\n def __init__(self,input_size,output_size,dropout=0.4):\n self.input_size = input_size\n self.output_size = output_size\n self.dropout = dropout\n self.generator = self.generator_network()\n self.discriminator = self.discriminator_network()\n self.adverserial = self.adverserial_network()\n \n def discriminator_trainable(self, val):\n self.discriminator.trainable = val\n for l in self.discriminator.layers:\n l.trainable = val\n \n def generator_network(self): # Generator : Object(6) - Dense - Object(4)\n self.g_input = Input(shape=(self.input_size,), name=\"Generator_Input\")\n g = Dense(128, activation='relu')(self.g_input)\n g = Dropout(self.dropout)(g)\n g = Dense(256, activation='relu')(g)\n g = Dropout(self.dropout)(g)\n g = Dense(512, activation='relu')(g)\n g = Dropout(self.dropout)(g)\n g = Dense(256, activation='relu')(g)\n g = Dropout(self.dropout)(g)\n g = Dense(128, activation='relu')(self.g_input)\n g = Dropout(self.dropout)(g)\n self.g_output = Dense(self.output_size, activation='tanh', name=\"Generator_Output\")(g) \n m = Model(self.g_input, self.g_output, name=\"Generator\")\n return m\n \n def discriminator_network(self): # Discriminator : Object(10) - Dense - Probability\n d_opt = RMSprop(lr=0.000125,decay=6e-8)\n d_input = Input(shape=(self.input_size+self.output_size,), name=\"Discriminator_Input\")\n d = Dense(128, activation='relu')(d_input)\n d = Dense(256, activation='relu')(d)\n d = Dense(512, activation='relu')(d)\n d = Dense(256, activation='relu')(d)\n d = Dense(128, activation='relu')(d)\n d_output = Dense(1, activation='sigmoid', name=\"Discriminator_Output\")(d)\n m = Model(d_input, d_output, name=\"Discriminator\")\n m.compile(loss='binary_crossentropy', optimizer=d_opt)\n return m\n \n def adverserial_network(self): # Adverserial : Object(6) - Generator - Discriminator - Probability\n a_opt = RMSprop(lr=0.0001,decay=3e-8)\n d_input = Concatenate(name=\"Generator_Input_Output\")([self.g_input,self.g_output])\n m=Model(self.g_input, self.discriminator(d_input))\n m.compile(loss='binary_crossentropy', optimizer=a_opt)\n return m\n \n def train_discriminator(self,val):\n self.discriminator.trainable = val\n for l in self.discriminator.layers:\n l.trainable = val\n \n def train(self, adverserial_set, discriminator_set, epochs, batch_size):\n \n losses = {\"d\":[], \"g\":[]}\n\n for i in range(epochs):\n \n batch = discriminator_set[int(i/2*batch_size/2):int((i/2+1)*batch_size/2)] # Gets a batch of real data\n \n for j in adverserial_set[int(i/2*batch_size/2):int((i/2+1)*batch_size/2)]: # Gets a batch of generated data\n n = copy.deepcopy(j)\n p = self.predict(j)\n for e in p:\n n.append(e)\n batch.append(n)\n \n #self.train_discriminator(True) # Turns on discriminator weights\n \n output = np.zeros(batch_size) # Sets output weight 0 for real and 1 for fakes\n output[int(batch_size/2):] = 1\n \n losses[\"d\"].append(self.discriminator.train_on_batch(np.array(batch), np.array(output))) # Train discriminator\n \n batch = adverserial_set[(i*batch_size):((i+1)*batch_size)] # Gets real data to train generator\n output = np.zeros(batch_size)\n \n #self.train_discriminator(False) # Turns off discriminator weights\n \n losses[\"g\"].append(self.adverserial.train_on_batch(np.array(batch), np.array(output))) # Train generator\n \n print('Epoch %s - Adverserial Loss : %s, Discriminator Loss : %s' % (i+1, losses[\"g\"][-1], losses[\"d\"][-1]))\n \n self.generator.save(\"Generator.h5\")\n self.discriminator.save(\"Discriminator.h5\")\n \n return losses\n \n def predict(self, pred):\n return self.generator.predict(np.array(pred).reshape(-1,6))[0]",
"_____no_output_____"
]
],
[
[
"# Training Data",
"_____no_output_____"
]
],
[
[
"training_set = []\nactual_set = []\n\nfor i in range(number_of_particles):\n p = Particle()\n if(i%2==0):\n training_set.append(p.get_list())\n else:\n actual_set.append(p.get_list_physics(dt))",
"_____no_output_____"
]
],
[
[
"# Training",
"_____no_output_____"
]
],
[
[
"network = GAN(input_size=6,output_size=4,dropout=0)",
"_____no_output_____"
],
[
"loss = network.train(adverserial_set=training_set,discriminator_set=actual_set,epochs=epochs,batch_size=batch_size)",
"Epoch 1 - Adverserial Loss : 0.68993413, Discriminator Loss : 0.6928395\nEpoch 2 - Adverserial Loss : 0.66143435, Discriminator Loss : 0.68966395\nEpoch 3 - Adverserial Loss : 0.6506674, Discriminator Loss : 0.69156027\nEpoch 4 - Adverserial Loss : 0.6345487, Discriminator Loss : 0.6851229\nEpoch 5 - Adverserial Loss : 0.6190306, Discriminator Loss : 0.6815617\nEpoch 6 - Adverserial Loss : 0.60411817, Discriminator Loss : 0.67499703\nEpoch 7 - Adverserial Loss : 0.5817479, Discriminator Loss : 0.6683197\nEpoch 8 - Adverserial Loss : 0.56560206, Discriminator Loss : 0.6666347\nEpoch 9 - Adverserial Loss : 0.5741377, Discriminator Loss : 0.68710524\nEpoch 10 - Adverserial Loss : 0.54610217, Discriminator Loss : 0.672557\nEpoch 11 - Adverserial Loss : 0.5515704, Discriminator Loss : 0.6653353\nEpoch 12 - Adverserial Loss : 0.5424552, Discriminator Loss : 0.6867492\nEpoch 13 - Adverserial Loss : 0.52841425, Discriminator Loss : 0.6719169\nEpoch 14 - Adverserial Loss : 0.52467513, Discriminator Loss : 0.66271853\nEpoch 15 - Adverserial Loss : 0.5150865, Discriminator Loss : 0.6709579\nEpoch 16 - Adverserial Loss : 0.49996227, Discriminator Loss : 0.67780423\nEpoch 17 - Adverserial Loss : 0.50212705, Discriminator Loss : 0.696942\nEpoch 18 - Adverserial Loss : 0.49521327, Discriminator Loss : 0.6863136\nEpoch 19 - Adverserial Loss : 0.48128614, Discriminator Loss : 0.67034364\nEpoch 20 - Adverserial Loss : 0.5052789, Discriminator Loss : 0.69045603\nEpoch 21 - Adverserial Loss : 0.49123746, Discriminator Loss : 0.6820578\nEpoch 22 - Adverserial Loss : 0.4826761, Discriminator Loss : 0.67253244\nEpoch 23 - Adverserial Loss : 0.47455353, Discriminator Loss : 0.64880645\nEpoch 24 - Adverserial Loss : 0.4836581, Discriminator Loss : 0.66169447\nEpoch 25 - Adverserial Loss : 0.469598, Discriminator Loss : 0.6742211\nEpoch 26 - Adverserial Loss : 0.4893468, Discriminator Loss : 0.6515214\nEpoch 27 - Adverserial Loss : 0.4918999, Discriminator Loss : 0.67179525\nEpoch 28 - Adverserial Loss : 0.48232928, Discriminator Loss : 0.68452317\nEpoch 29 - Adverserial Loss : 0.46680003, Discriminator Loss : 0.649647\nEpoch 30 - Adverserial Loss : 0.46584213, Discriminator Loss : 0.6481444\nEpoch 31 - Adverserial Loss : 0.44775116, Discriminator Loss : 0.69185436\nEpoch 32 - Adverserial Loss : 0.45568126, Discriminator Loss : 0.69919485\nEpoch 33 - Adverserial Loss : 0.46387553, Discriminator Loss : 0.67933834\nEpoch 34 - Adverserial Loss : 0.4780157, Discriminator Loss : 0.65117264\nEpoch 35 - Adverserial Loss : 0.4502793, Discriminator Loss : 0.64282584\nEpoch 36 - Adverserial Loss : 0.46630493, Discriminator Loss : 0.6785953\nEpoch 37 - Adverserial Loss : 0.45504636, Discriminator Loss : 0.70681316\nEpoch 38 - Adverserial Loss : 0.45864415, Discriminator Loss : 0.69120395\nEpoch 39 - Adverserial Loss : 0.46821624, Discriminator Loss : 0.68380153\nEpoch 40 - Adverserial Loss : 0.463122, Discriminator Loss : 0.68495214\nEpoch 41 - Adverserial Loss : 0.48433882, Discriminator Loss : 0.6410985\nEpoch 42 - Adverserial Loss : 0.46019632, Discriminator Loss : 0.6659787\nEpoch 43 - Adverserial Loss : 0.4740353, Discriminator Loss : 0.68378663\nEpoch 44 - Adverserial Loss : 0.44774973, Discriminator Loss : 0.6546668\nEpoch 45 - Adverserial Loss : 0.47461084, Discriminator Loss : 0.6623768\nEpoch 46 - Adverserial Loss : 0.4374383, Discriminator Loss : 0.6582193\nEpoch 47 - Adverserial Loss : 0.44694984, Discriminator Loss : 0.67210805\nEpoch 48 - Adverserial Loss : 0.46363854, Discriminator Loss : 0.6862819\nEpoch 49 - Adverserial Loss : 0.45307237, Discriminator Loss : 0.63064533\nEpoch 50 - Adverserial Loss : 0.43084458, Discriminator Loss : 0.61385685\nEpoch 51 - Adverserial Loss : 0.45059398, Discriminator Loss : 0.6510699\nEpoch 52 - Adverserial Loss : 0.4427229, Discriminator Loss : 0.6816915\nEpoch 53 - Adverserial Loss : 0.43752646, Discriminator Loss : 0.6853812\nEpoch 54 - Adverserial Loss : 0.4232367, Discriminator Loss : 0.6900676\nEpoch 55 - Adverserial Loss : 0.4301697, Discriminator Loss : 0.6708988\nEpoch 56 - Adverserial Loss : 0.45538002, Discriminator Loss : 0.69781375\nEpoch 57 - Adverserial Loss : 0.44145784, Discriminator Loss : 0.7204728\nEpoch 58 - Adverserial Loss : 0.44863936, Discriminator Loss : 0.67540836\nEpoch 59 - Adverserial Loss : 0.43241632, Discriminator Loss : 0.6707108\nEpoch 60 - Adverserial Loss : 0.4435634, Discriminator Loss : 0.67677605\nEpoch 61 - Adverserial Loss : 0.45064044, Discriminator Loss : 0.7325363\nEpoch 62 - Adverserial Loss : 0.45267245, Discriminator Loss : 0.7074709\nEpoch 63 - Adverserial Loss : 0.45730782, Discriminator Loss : 0.64637834\nEpoch 64 - Adverserial Loss : 0.4440861, Discriminator Loss : 0.6755284\nEpoch 65 - Adverserial Loss : 0.45689663, Discriminator Loss : 0.65478253\nEpoch 66 - Adverserial Loss : 0.43381196, Discriminator Loss : 0.61726266\nEpoch 67 - Adverserial Loss : 0.45423836, Discriminator Loss : 0.682735\nEpoch 68 - Adverserial Loss : 0.44567546, Discriminator Loss : 0.6633605\nEpoch 69 - Adverserial Loss : 0.46208915, Discriminator Loss : 0.6470846\nEpoch 70 - Adverserial Loss : 0.43579426, Discriminator Loss : 0.67914534\nEpoch 71 - Adverserial Loss : 0.44100276, Discriminator Loss : 0.7011275\nEpoch 72 - Adverserial Loss : 0.4595195, Discriminator Loss : 0.72181153\nEpoch 73 - Adverserial Loss : 0.4372614, Discriminator Loss : 0.68018055\nEpoch 74 - Adverserial Loss : 0.4659946, Discriminator Loss : 0.67874914\nEpoch 75 - Adverserial Loss : 0.43561178, Discriminator Loss : 0.71172446\nEpoch 76 - Adverserial Loss : 0.4276686, Discriminator Loss : 0.6545134\nEpoch 77 - Adverserial Loss : 0.42894298, Discriminator Loss : 0.6359602\nEpoch 78 - Adverserial Loss : 0.4413724, Discriminator Loss : 0.681329\nEpoch 79 - Adverserial Loss : 0.46897948, Discriminator Loss : 0.68444234\nEpoch 80 - Adverserial Loss : 0.4456116, Discriminator Loss : 0.6681315\nEpoch 81 - Adverserial Loss : 0.4755069, Discriminator Loss : 0.6676806\nEpoch 82 - Adverserial Loss : 0.42014265, Discriminator Loss : 0.6799015\nEpoch 83 - Adverserial Loss : 0.47746795, Discriminator Loss : 0.67737925\nEpoch 84 - Adverserial Loss : 0.43996295, Discriminator Loss : 0.67604256\nEpoch 85 - Adverserial Loss : 0.4327209, Discriminator Loss : 0.6708926\nEpoch 86 - Adverserial Loss : 0.44610336, Discriminator Loss : 0.66637367\nEpoch 87 - Adverserial Loss : 0.44142282, Discriminator Loss : 0.68081385\nEpoch 88 - Adverserial Loss : 0.45937046, Discriminator Loss : 0.73096496\nEpoch 89 - Adverserial Loss : 0.43894643, Discriminator Loss : 0.6767824\nEpoch 90 - Adverserial Loss : 0.4451853, Discriminator Loss : 0.6382792\nEpoch 91 - Adverserial Loss : 0.44680282, Discriminator Loss : 0.69345856\nEpoch 92 - Adverserial Loss : 0.41137543, Discriminator Loss : 0.69330645\nEpoch 93 - Adverserial Loss : 0.4686013, Discriminator Loss : 0.68496954\nEpoch 94 - Adverserial Loss : 0.47020942, Discriminator Loss : 0.68736047\nEpoch 95 - Adverserial Loss : 0.44107744, Discriminator Loss : 0.63904953\nEpoch 96 - Adverserial Loss : 0.43405592, Discriminator Loss : 0.6586726\nEpoch 97 - Adverserial Loss : 0.47728264, Discriminator Loss : 0.68063956\nEpoch 98 - Adverserial Loss : 0.45187652, Discriminator Loss : 0.6572933\nEpoch 99 - Adverserial Loss : 0.44229847, Discriminator Loss : 0.6528614\nEpoch 100 - Adverserial Loss : 0.4581712, Discriminator Loss : 0.6587184\nEpoch 101 - Adverserial Loss : 0.4100021, Discriminator Loss : 0.6640239\nEpoch 102 - Adverserial Loss : 0.44911265, Discriminator Loss : 0.64451975\nEpoch 103 - Adverserial Loss : 0.44372746, Discriminator Loss : 0.6111923\nEpoch 104 - Adverserial Loss : 0.42413574, Discriminator Loss : 0.6077135\nEpoch 105 - Adverserial Loss : 0.38416517, Discriminator Loss : 0.63506794\nEpoch 106 - Adverserial Loss : 0.44143242, Discriminator Loss : 0.647977\nEpoch 107 - Adverserial Loss : 0.45337635, Discriminator Loss : 0.6554244\nEpoch 108 - Adverserial Loss : 0.44754887, Discriminator Loss : 0.6385607\nEpoch 109 - Adverserial Loss : 0.44343334, Discriminator Loss : 0.63654506\nEpoch 110 - Adverserial Loss : 0.4174673, Discriminator Loss : 0.63702285\nEpoch 111 - Adverserial Loss : 0.4576087, Discriminator Loss : 0.6328316\nEpoch 112 - Adverserial Loss : 0.41836363, Discriminator Loss : 0.64255047\nEpoch 113 - Adverserial Loss : 0.45875356, Discriminator Loss : 0.6508331\nEpoch 114 - Adverserial Loss : 0.47654557, Discriminator Loss : 0.64116377\nEpoch 115 - Adverserial Loss : 0.44722623, Discriminator Loss : 0.6492766\nEpoch 116 - Adverserial Loss : 0.43521494, Discriminator Loss : 0.6263341\n"
],
[
"fig = plt.figure(figsize=(13,7))\nplt.title(\"Loss Function over Epochs\")\nplt.xlabel(\"Epoch\")\nplt.ylabel(\"Loss\")\nplt.plot(loss[\"g\"], label=\"Adversarial Loss\")\nplt.plot(loss[\"d\"], label=\"Discriminative Loss\")\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"network.predict([0.1,0.2,0.1,0.1,0.1,0.1])",
"_____no_output_____"
],
[
"network.generator.summary()\nnetwork.discriminator.summary()\nnetwork.adverserial.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nGenerator_Input (InputLayer) (None, 6) 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 128) 896 \n_________________________________________________________________\ndropout_4 (Dropout) (None, 128) 0 \n_________________________________________________________________\nGenerator_Output (Dense) (None, 4) 516 \n=================================================================\nTotal params: 1,412\nTrainable params: 1,412\nNon-trainable params: 0\n_________________________________________________________________\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nDiscriminator_Input (InputLa (None, 10) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 128) 1408 \n_________________________________________________________________\ndense_6 (Dense) (None, 256) 33024 \n_________________________________________________________________\ndense_7 (Dense) (None, 512) 131584 \n_________________________________________________________________\ndense_8 (Dense) (None, 256) 131328 \n_________________________________________________________________\ndense_9 (Dense) (None, 128) 32896 \n_________________________________________________________________\nDiscriminator_Output (Dense) (None, 1) 129 \n=================================================================\nTotal params: 330,369\nTrainable params: 330,369\nNon-trainable params: 0\n_________________________________________________________________\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nGenerator_Input (InputLayer) (None, 6) 0 \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 128) 896 Generator_Input[0][0] \n__________________________________________________________________________________________________\ndropout_4 (Dropout) (None, 128) 0 dense_4[0][0] \n__________________________________________________________________________________________________\nGenerator_Output (Dense) (None, 4) 516 dropout_4[0][0] \n__________________________________________________________________________________________________\nGenerator_Input_Output (Concate (None, 10) 0 Generator_Input[0][0] \n Generator_Output[0][0] \n__________________________________________________________________________________________________\nDiscriminator (Model) (None, 1) 330369 Generator_Input_Output[0][0] \n==================================================================================================\nTotal params: 331,781\nTrainable params: 331,781\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0b7a87c2c5b6dd4b92eaea08ac1c343fe35f664 | 44,941 | ipynb | Jupyter Notebook | notebooks/TightBinding Tests.ipynb | Quejiahao/SKTB.jl | aee9f9fd5beccfcb36120a0fac4b977304e52a49 | [
"MIT"
] | 5 | 2016-09-11T16:47:47.000Z | 2019-06-29T09:53:23.000Z | notebooks/TightBinding Tests.ipynb | Quejiahao/SKTB.jl | aee9f9fd5beccfcb36120a0fac4b977304e52a49 | [
"MIT"
] | 22 | 2016-08-23T18:25:39.000Z | 2019-06-30T12:50:33.000Z | notebooks/TightBinding Tests.ipynb | Quejiahao/SKTB.jl | aee9f9fd5beccfcb36120a0fac4b977304e52a49 | [
"MIT"
] | 2 | 2018-04-27T15:15:44.000Z | 2018-11-27T21:39:17.000Z | 27.08921 | 104 | 0.408714 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b7aa88575802b5c04cbf6f6248b50dec55dd1d | 18,252 | ipynb | Jupyter Notebook | docs/field-operations.ipynb | ubermag/discretisedfield | fec016c85fcc091006e678845bca999b993b987c | [
"BSD-3-Clause"
] | 9 | 2019-08-30T14:00:43.000Z | 2022-01-16T15:01:44.000Z | docs/field-operations.ipynb | ubermag/discretisedfield | fec016c85fcc091006e678845bca999b993b987c | [
"BSD-3-Clause"
] | 50 | 2019-06-13T13:41:57.000Z | 2022-03-28T09:14:33.000Z | docs/field-operations.ipynb | ubermag/discretisedfield | fec016c85fcc091006e678845bca999b993b987c | [
"BSD-3-Clause"
] | 7 | 2019-08-28T14:16:10.000Z | 2021-12-13T21:06:06.000Z | 21.397421 | 408 | 0.502192 | [
[
[
"# Field operations\n\nThere are several convenience methods that can be used to analyse the field. Let us first define the mesh we are going to work with.",
"_____no_output_____"
]
],
[
[
"import discretisedfield as df\n\np1 = (-50, -50, -50)\np2 = (50, 50, 50)\nn = (2, 2, 2)\nmesh = df.Mesh(p1=p1, p2=p2, n=n)",
"_____no_output_____"
]
],
[
[
"We are going to initialise the vector field (`dim=3`), with\n\n$$\\mathbf{f}(x, y, z) = (xy, 2xy, xyz)$$\n\nFor that, we are going to use the following Python function.",
"_____no_output_____"
]
],
[
[
"def value_function(pos):\n x, y, z = pos\n return x*y, 2*x*y, x*y*z",
"_____no_output_____"
]
],
[
[
"Finally, our field is",
"_____no_output_____"
]
],
[
[
"field = df.Field(mesh, dim=3, value=value_function)",
"_____no_output_____"
]
],
[
[
"## 1. Sampling the field\n\nAs we have shown previously, a field can be sampled by calling it. The argument must be a 3-length iterable and it contains the coordinates of the point.",
"_____no_output_____"
]
],
[
[
"point = (0, 0, 0)\nfield(point)",
"_____no_output_____"
]
],
[
[
"However if the point is outside the mesh, an exception is raised.",
"_____no_output_____"
]
],
[
[
"point = (100, 100, 100)\ntry:\n field(point)\nexcept ValueError:\n print('Exception raised.')",
"Exception raised.\n"
]
],
[
[
"## 2. Extracting the component of a vector field\n\nA three-dimensional vector field can be understood as three separate scalar fields, where each scalar field is a component of a vector field value. A scalar field of a component can be extracted by accessing `x`, `y`, or `z` attribute of the field.",
"_____no_output_____"
]
],
[
[
"x_component = field.x\nx_component((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"Default names `x`, `y`, and (for dim 3) `z` are only available for fields with dimensionality 2 or 3.",
"_____no_output_____"
]
],
[
[
"field.components",
"_____no_output_____"
]
],
[
[
"It is possible to change the component names:",
"_____no_output_____"
]
],
[
[
"field.components = ['mx', 'my', 'mz']\nfield.mx((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"This overrides the component labels and the old `x`, `y` and `z` cannot be used anymore:",
"_____no_output_____"
]
],
[
[
"try:\n field.x\nexcept AttributeError as e:\n print(e)",
"Object has no attribute x.\n"
]
],
[
[
"We change the component labels back to `x`, `y`, and `z` for the rest of this notebook.",
"_____no_output_____"
]
],
[
[
"field.components = ['x', 'y', 'z']",
"_____no_output_____"
]
],
[
[
"Custom component names can optionally also be specified during field creation. If not specified, the default values are used for fields with dimensions 2 or 3. Higher-dimensional fields have no defaults and custom labes have to be specified in order to access individual field components:",
"_____no_output_____"
]
],
[
[
"field_4d = df.Field(mesh, dim=4, value=[1, 1, 1, 1], components=['c1', 'c2', 'c3', 'c4'])\nfield_4d",
"_____no_output_____"
],
[
"field_4d.c1((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"## 3. Computing the average\n\nThe average of the field can be obtained by calling `discretisedfield.Field.average` property.",
"_____no_output_____"
]
],
[
[
"field.average",
"_____no_output_____"
]
],
[
[
"Average always return a tuple, independent of the dimension of the field's value.",
"_____no_output_____"
]
],
[
[
"field.x.average",
"_____no_output_____"
]
],
[
[
"## 4. Iterating through the field\n\nThe field object itself is an iterable. That means that it can be iterated through. As a result it returns a tuple, where the first element is the coordinate of the mesh point, whereas the second one is its value.",
"_____no_output_____"
]
],
[
[
"for coordinate, value in field:\n print(coordinate, value)",
"(-25.0, -25.0, -25.0) (625.0, 1250.0, -15625.0)\n(25.0, -25.0, -25.0) (-625.0, -1250.0, 15625.0)\n(-25.0, 25.0, -25.0) (-625.0, -1250.0, 15625.0)\n(25.0, 25.0, -25.0) (625.0, 1250.0, -15625.0)\n(-25.0, -25.0, 25.0) (625.0, 1250.0, 15625.0)\n(25.0, -25.0, 25.0) (-625.0, -1250.0, -15625.0)\n(-25.0, 25.0, 25.0) (-625.0, -1250.0, -15625.0)\n(25.0, 25.0, 25.0) (625.0, 1250.0, 15625.0)\n"
]
],
[
[
"## 5. Sampling the field along the line\n\nTo sample the points of the field which are on a certain line, `discretisedfield.Field.line` method is used. It takes two points `p1` and `p2` that define the line and an integer `n` which defines how many mesh coordinates on that line are required. The default value of `n` is 100.",
"_____no_output_____"
]
],
[
[
"line = field.line(p1=(-10, 0, 0), p2=(10, 0, 0), n=5)",
"_____no_output_____"
]
],
[
[
"## 6. Intersecting the field with a plane\n\nIf we intersect the field with a plane, `discretisedfield.Field.plane` will return a new field object which contains only discretisation cells that belong to that plane. The planes allowed are the planes perpendicular to the axes of the Cartesian coordinate system. For instance, a plane parallel to the $yz$-plane (perpendicular to the $x$-axis) which intesects the $x$-axis at 1, can be written as\n\n$$x = 1$$",
"_____no_output_____"
]
],
[
[
"field.plane(x=1)",
"_____no_output_____"
]
],
[
[
"If we want to cut through the middle of the mesh, we do not need to provide a particular value for a coordinate.",
"_____no_output_____"
]
],
[
[
"field.plane('x')",
"_____no_output_____"
]
],
[
[
"## 7. Cascading the operations\n\nLet us say we want to compute the average of an $x$ component of the field on the plane $y=10$. In order to do that, we can cascade several operation in a single line.",
"_____no_output_____"
]
],
[
[
"field.plane(y=10).x.average",
"_____no_output_____"
]
],
[
[
"This gives the same result as for instance",
"_____no_output_____"
]
],
[
[
"field.x.plane(y=10).average",
"_____no_output_____"
]
],
[
[
"## 8. Complex fields\n\n`discretisedfield` supports complex-valued fields.",
"_____no_output_____"
]
],
[
[
"cfield = df.Field(mesh, dim=3, value=(1+1.5j, 2, 3j))",
"_____no_output_____"
]
],
[
[
"We can extract `real` and `imaginary` part.",
"_____no_output_____"
]
],
[
[
"cfield.real((0, 0, 0))",
"_____no_output_____"
],
[
"cfield.imag((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"Similarly we get `real` and `imaginary` parts of individual components.",
"_____no_output_____"
]
],
[
[
"cfield.x.real((0, 0, 0))",
"_____no_output_____"
],
[
"cfield.x.imag((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"Complex conjugate.",
"_____no_output_____"
]
],
[
[
"cfield.conjugate((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"Phase in the complex plane.",
"_____no_output_____"
]
],
[
[
"cfield.phase((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"## 9. Applying `numpys` universal functions\nAll numpy universal functions can be applied to `discretisedfield.Field` objects. Below we show a different examples. For available functions please refer to the `numpy` [documentation](https://numpy.org/doc/stable/reference/ufuncs.html#available-ufuncs).",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"f1 = df.Field(mesh, dim=1, value=1)\nf2 = df.Field(mesh, dim=1, value=np.pi)\nf3 = df.Field(mesh, dim=1, value=2)",
"_____no_output_____"
],
[
"np.sin(f1)",
"_____no_output_____"
],
[
"np.sin(f2)((0, 0, 0))",
"_____no_output_____"
],
[
"np.sum((f1, f2, f3))((0, 0, 0))",
"_____no_output_____"
],
[
"np.exp(f1)((0, 0, 0))",
"_____no_output_____"
],
[
"np.power(f3, 2)((0, 0, 0))",
"_____no_output_____"
]
],
[
[
"## Other\n\nFull description of all existing functionality can be found in the [API Reference](https://discretisedfield.readthedocs.io/en/latest/_autosummary/discretisedfield.Field.html).",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
d0b7ba0cff14f09b6d6d59bf1a7e22b7889ba835 | 16,807 | ipynb | Jupyter Notebook | 01_Getting_&_Knowing_Your_Data/World Food Facts/Exercises.ipynb | GregoireCourtois/pandas_exercises | 6cab3ed818181e2b162b6d17a0cba2f11e225e8e | [
"BSD-3-Clause"
] | null | null | null | 01_Getting_&_Knowing_Your_Data/World Food Facts/Exercises.ipynb | GregoireCourtois/pandas_exercises | 6cab3ed818181e2b162b6d17a0cba2f11e225e8e | [
"BSD-3-Clause"
] | null | null | null | 01_Getting_&_Knowing_Your_Data/World Food Facts/Exercises.ipynb | GregoireCourtois/pandas_exercises | 6cab3ed818181e2b162b6d17a0cba2f11e225e8e | [
"BSD-3-Clause"
] | null | null | null | 28.680887 | 240 | 0.415601 | [
[
[
"# Exercise 1",
"_____no_output_____"
],
[
"### Step 1. Go to https://www.kaggle.com/openfoodfacts/world-food-facts/data",
"_____no_output_____"
],
[
"### Step 2. Download the dataset to your computer and unzip it.",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### Step 3. Use the tsv file and assign it to a dataframe called food",
"_____no_output_____"
]
],
[
[
"Location = r'/Users/GregoireCourtois/Desktop/Python_programmes/Pandas exercises/pandas_exercises-master/01_Getting_&_Knowing_Your_Data/World Food Facts/en.openfoodfacts.org.products.tsv'\nfood = pd.read_csv(Location, sep='\\t') #tsv file are tabular spaced! csv fil are comma spaced",
"/Applications/anaconda3/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3146: DtypeWarning: Columns (0,3,5,19,20,24,25,26,27,28,36,37,38,39,48) have mixed types.Specify dtype option on import or set low_memory=False.\n has_raised = await self.run_ast_nodes(code_ast.body, cell_name,\n"
]
],
[
[
"### Step 4. See the first 5 entries",
"_____no_output_____"
]
],
[
[
"food.head()",
"_____no_output_____"
]
],
[
[
"### Step 5. What is the number of observations in the dataset?",
"_____no_output_____"
]
],
[
[
"food.shape",
"_____no_output_____"
],
[
"food.shape[0]",
"_____no_output_____"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"food.shape[1]",
"_____no_output_____"
],
[
"food.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 356027 entries, 0 to 356026\nColumns: 163 entries, code to water-hardness_100g\ndtypes: float64(107), object(56)\nmemory usage: 442.8+ MB\n"
]
],
[
[
"### Step 7. Print the name of all the columns.",
"_____no_output_____"
]
],
[
[
"food.columns",
"_____no_output_____"
]
],
[
[
"### Step 8. What is the name of 105th column?",
"_____no_output_____"
]
],
[
[
"food.columns[104]",
"_____no_output_____"
]
],
[
[
"### Step 9. What is the type of the observations of the 105th column?",
"_____no_output_____"
]
],
[
[
"food['-glucose_100g'].dtype",
"_____no_output_____"
]
],
[
[
"### Step 10. How is the dataset indexed?",
"_____no_output_____"
]
],
[
[
"food.index",
"_____no_output_____"
]
],
[
[
"### Step 11. What is the product name of the 19th observation?",
"_____no_output_____"
]
],
[
[
"food['product_name'][18]",
"_____no_output_____"
],
[
"food.values[18][7] #way longer",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0b7ba4aa6e455ed051e726b6f3f31525adfc579 | 12,352 | ipynb | Jupyter Notebook | source/02-portfolios-tutorials/notebook/Returns On Single Assets.ipynb | EmanuelFontelles/machineLearningForTrading | c7741f52bc11fe362ff7455c678724d142043f1d | [
"MIT"
] | null | null | null | source/02-portfolios-tutorials/notebook/Returns On Single Assets.ipynb | EmanuelFontelles/machineLearningForTrading | c7741f52bc11fe362ff7455c678724d142043f1d | [
"MIT"
] | null | null | null | source/02-portfolios-tutorials/notebook/Returns On Single Assets.ipynb | EmanuelFontelles/machineLearningForTrading | c7741f52bc11fe362ff7455c678724d142043f1d | [
"MIT"
] | null | null | null | 34.216066 | 1,187 | 0.473931 | [
[
[
"# **Let's check out the calculation of rates of returns on single assets**\n\nThe steps are as follows;\n\n1. Import Python's number crunching libraries\n2. Use panda's data reader to get real world stock information of Apple\n3. Explore the data\n4. Calculate rate of return using the simple returns fomula \n5. Calculate rate of return using log returns\n\n**More info on Pandas Datareader:** https://pandas-datareader.readthedocs.io/en/latest/remote_data.html#\n\n** Documentation for .shift() method:** https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.shift.html\n\n**More info on the differences between simple returns and log returns:** https://quant.stackexchange.com/questions/4160/discrete-returns-versus-log-returns-of-assets",
"_____no_output_____"
]
],
[
[
"# Step 1 (import python's number crunchers)\n\nimport pandas as pd\nimport numpy as np\nfrom pandas_datareader import data as web",
"_____no_output_____"
],
[
"# Step 2 & 3 (Get Apple stock information using Pandas Datareader)\n\ndata = pd.DataFrame()\n\ntickers = ['AAPL']\n\nfor item in tickers:\n data[item] = web.DataReader(item, data_source='yahoo', start='01-01-2000')['Adj Close']\n\ndata.head()",
"_____no_output_____"
],
[
"# Step 4 (Simple Returns with the formula)\n# .shift() method to use previous value \n\nsimple_returns1 = (data / data.shift(1)) - 1\nsimple_returns1.head()",
"_____no_output_____"
],
[
"# Still Step 4 (Simple Returns formula expressed as a method)\n# Same result as above\n# Alternative solution\n\nsimple_returns2 = data.pct_change()\nsimple_returns2.head()",
"_____no_output_____"
],
[
"# Step 5 (Getting log returns)\n\nlog_returns = np.log(data / data.shift(1))\nlog_returns.head()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0b7bab8638ba311e5eb5c6588bf00605131aa28 | 4,740 | ipynb | Jupyter Notebook | src/Floatpoint.ipynb | hessianguo/NumericalMethod.jl | bd6c00a88c8168e39b2ba1894466a6b6f6e24984 | [
"MIT"
] | null | null | null | src/Floatpoint.ipynb | hessianguo/NumericalMethod.jl | bd6c00a88c8168e39b2ba1894466a6b6f6e24984 | [
"MIT"
] | null | null | null | src/Floatpoint.ipynb | hessianguo/NumericalMethod.jl | bd6c00a88c8168e39b2ba1894466a6b6f6e24984 | [
"MIT"
] | null | null | null | 16.928571 | 77 | 0.477004 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
d0b7c0ca7b0c156bcdb42346480af7ca6beb3497 | 2,571 | ipynb | Jupyter Notebook | Classifying Sentiment of Restaurant Reviews/download-data.ipynb | JKooll/Cookbook | 6431bb0a6bdee431ff150891bbfeb368d2c29479 | [
"MIT"
] | null | null | null | Classifying Sentiment of Restaurant Reviews/download-data.ipynb | JKooll/Cookbook | 6431bb0a6bdee431ff150891bbfeb368d2c29479 | [
"MIT"
] | null | null | null | Classifying Sentiment of Restaurant Reviews/download-data.ipynb | JKooll/Cookbook | 6431bb0a6bdee431ff150891bbfeb368d2c29479 | [
"MIT"
] | null | null | null | 30.247059 | 110 | 0.536367 | [
[
[
"import requests\nimport os\nfrom pathlib import Path\n\ndef progress_bar(some_iter):\n try:\n from tqdm import tqdm\n return tqdm(some_iter)\n except ModuleNotFoundError:\n return some_iter\n\ndef download_file_from_google_drive(id, destination):\n print(\"Trying to fetch {}\".format(destination))\n\n def get_confirm_token(response):\n for key, value in response.cookies.items():\n if key.startswith('download_warning'):\n return value\n\n return None\n\n def save_response_content(response, destination):\n CHUNK_SIZE = 32768\n\n if not os.path.exists(os.path.split(destination)[0]):\n path = Path(os.path.split(destination)[0])\n path.mkdir(parents=True)\n\n with open(destination, \"wb\") as f:\n for chunk in progress_bar(response.iter_content(CHUNK_SIZE)):\n if chunk: # filter out keep-alive new chunks\n f.write(chunk)\n\n URL = \"https://docs.google.com/uc?export=download\"\n\n session = requests.Session()\n\n response = session.get(URL, params = { 'id' : id }, stream = True)\n token = get_confirm_token(response)\n\n if token:\n params = { 'id' : id, 'confirm' : token }\n response = session.get(URL, params = params, stream = True)\n\n save_response_content(response, destination)",
"_____no_output_____"
],
[
"download_file_from_google_drive(\"1xeUnqkhuzGGzZKThzPeXe2Vf6Uu_g_xM\", \"./data/yelp/raw_train.csv\")\ndownload_file_from_google_drive('1G42LXv72DrhK4QKJoFhabVL4IU6v2ZvB', './data/yelp/raw_test.csv')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
d0b7c15327a80dde96a0c81ddf161afe027feb4a | 134,075 | ipynb | Jupyter Notebook | CRIM_Intervals_Classify_12_31_20.ipynb | RichardFreedman/CRIM-notebooks | e2aa0b0798615898644df9fe0cc92fe532a4bbdc | [
"MIT"
] | null | null | null | CRIM_Intervals_Classify_12_31_20.ipynb | RichardFreedman/CRIM-notebooks | e2aa0b0798615898644df9fe0cc92fe532a4bbdc | [
"MIT"
] | null | null | null | CRIM_Intervals_Classify_12_31_20.ipynb | RichardFreedman/CRIM-notebooks | e2aa0b0798615898644df9fe0cc92fe532a4bbdc | [
"MIT"
] | null | null | null | 86.000641 | 21,738 | 0.426754 | [
[
[
"## Search with Options\n\n- Piece or Corpus\n- Actual or Incremental Durations\n- Chromatic or Diatonic\n- Exact or Close\n- Classify\n\n***\n",
"_____no_output_____"
]
],
[
[
"from crim_intervals import *\nimport pandas as pd\nimport ast\nimport matplotlib\nfrom itertools import tee, combinations",
"_____no_output_____"
]
],
[
[
"### The Complete Corpus",
"_____no_output_____"
]
],
[
[
"work_list = ['CRIM_Mass_0001_1.mei','CRIM_Mass_0001_2.mei','CRIM_Mass_0001_3.mei','CRIM_Mass_0001_4.mei','CRIM_Mass_0001_5.mei','CRIM_Mass_0002_1.mei','CRIM_Mass_0002_2.mei','CRIM_Mass_0002_3.mei','CRIM_Mass_0002_4.mei','CRIM_Mass_0002_5.mei','CRIM_Mass_0003_1.mei','CRIM_Mass_0003_2.mei','CRIM_Mass_0003_3.mei','CRIM_Mass_0003_4.mei','CRIM_Mass_0003_5.mei','CRIM_Mass_0004_1.mei','CRIM_Mass_0004_2.mei','CRIM_Mass_0004_3.mei','CRIM_Mass_0004_4.mei','CRIM_Mass_0004_5.mei','CRIM_Mass_0005_1.mei','CRIM_Mass_0005_2.mei','CRIM_Mass_0005_3.mei','CRIM_Mass_0005_4.mei','CRIM_Mass_0005_5.mei','CRIM_Mass_0006_1.mei','CRIM_Mass_0006_2.mei','CRIM_Mass_0006_3.mei','CRIM_Mass_0006_4.mei','CRIM_Mass_0006_5.mei','CRIM_Mass_0007_1.mei', 'CRIM_Mass_0007_2.mei', 'CRIM_Mass_0007_3.mei', 'CRIM_Mass_0007_4.mei', 'CRIM_Mass_0007_5.mei', 'CRIM_Mass_0008_1.mei', 'CRIM_Mass_0008_2.mei', 'CRIM_Mass_0008_3.mei', 'CRIM_Mass_0008_4.mei', 'CRIM_Mass_0008_5.mei', 'CRIM_Mass_0009_1.mei', 'CRIM_Mass_0009_2.mei', 'CRIM_Mass_0009_3.mei', 'CRIM_Mass_0009_4.mei', 'CRIM_Mass_0009_5.mei', 'CRIM_Mass_0010_1.mei', 'CRIM_Mass_0010_2.mei', 'CRIM_Mass_0010_3.mei', 'CRIM_Mass_0010_4.mei', 'CRIM_Mass_0010_5.mei', 'CRIM_Mass_0011_1.mei', 'CRIM_Mass_0011_2.mei', 'CRIM_Mass_0011_3.mei', 'CRIM_Mass_0011_4.mei', 'CRIM_Mass_0011_5.mei', 'CRIM_Mass_0012_1.mei', 'CRIM_Mass_0012_2.mei', 'CRIM_Mass_0012_3.mei', 'CRIM_Mass_0012_4.mei', 'CRIM_Mass_0012_5.mei', 'CRIM_Mass_0013_1.mei', 'CRIM_Mass_0013_2.mei', 'CRIM_Mass_0013_3.mei', 'CRIM_Mass_0013_4.mei', 'CRIM_Mass_0013_5.mei', 'CRIM_Mass_0014_1.mei', 'CRIM_Mass_0014_2.mei', 'CRIM_Mass_0014_3.mei', 'CRIM_Mass_0014_4.mei', 'CRIM_Mass_0014_5.mei', 'CRIM_Mass_0015_1.mei', 'CRIM_Mass_0015_2.mei', 'CRIM_Mass_0015_3.mei', 'CRIM_Mass_0015_4.mei', 'CRIM_Mass_0015_5.mei', 'CRIM_Mass_0016_1.mei', 'CRIM_Mass_0016_2.mei', 'CRIM_Mass_0016_3.mei', 'CRIM_Mass_0016_4.mei', 'CRIM_Mass_0016_5.mei', 'CRIM_Mass_0017_1.mei', 'CRIM_Mass_0017_2.mei', 'CRIM_Mass_0017_3.mei', 'CRIM_Mass_0017_4.mei', 'CRIM_Mass_0017_5.mei', 'CRIM_Mass_0018_1.mei', 'CRIM_Mass_0018_2.mei', 'CRIM_Mass_0018_3.mei', 'CRIM_Mass_0018_4.mei', 'CRIM_Mass_0018_5.mei', 'CRIM_Mass_0019_1.mei', 'CRIM_Mass_0019_2.mei', 'CRIM_Mass_0019_3.mei', 'CRIM_Mass_0019_4.mei', 'CRIM_Mass_0019_5.mei', 'CRIM_Mass_0020_1.mei', 'CRIM_Mass_0020_2.mei', 'CRIM_Mass_0020_3.mei', 'CRIM_Mass_0020_4.mei', 'CRIM_Mass_0020_5.mei', 'CRIM_Mass_0021_1.mei', 'CRIM_Mass_0021_2.mei', 'CRIM_Mass_0021_3.mei', 'CRIM_Mass_0021_4.mei', 'CRIM_Mass_0021_5.mei', 'CRIM_Mass_0022_2.mei', 'CRIM_Model_0001.mei', 'CRIM_Model_0008.mei', 'CRIM_Model_0009.mei', 'CRIM_Model_0010.mei', 'CRIM_Model_0011.mei', 'CRIM_Model_0012.mei', 'CRIM_Model_0013.mei', 'CRIM_Model_0014.mei', 'CRIM_Model_0015.mei', 'CRIM_Model_0016.mei', 'CRIM_Model_0017.mei', 'CRIM_Model_0019.mei', 'CRIM_Model_0020.mei', 'CRIM_Model_0021.mei', 'CRIM_Model_0023.mei', 'CRIM_Model_0025.mei', 'CRIM_Model_0026.mei',\n]",
"_____no_output_____"
]
],
[
[
"### Short Corpus",
"_____no_output_____"
]
],
[
[
"work_list = ['CRIM_Mass_0002_1.mei',\n 'CRIM_Mass_0002_2.mei',\n 'CRIM_Mass_0002_3.mei',\n 'CRIM_Mass_0002_4.mei',\n 'CRIM_Mass_0002_5.mei',\n'CRIM_Model_0001.mei']\n\n# work_list = [\n# 'CRIM_Model_0008.mei']",
"_____no_output_____"
]
],
[
[
"## Load File and Correct the MEI Metadata",
"_____no_output_____"
]
],
[
[
"work_list = [el.replace(\"CRIM_\", \"https://crimproject.org/mei/MEI_4.0/CRIM_\") for el in work_list]\ncorpus = CorpusBase(work_list)\n\nimport xml.etree.ElementTree as ET\nimport requests\n\nMEINSURI = 'http://www.music-encoding.org/ns/mei'\nMEINS = '{%s}' % MEINSURI\n\nfor i, path in enumerate(work_list):\n \n try:\n if path[0] == '/':\n mei_doc = ET.parse(path)\n else:\n mei_doc = ET.fromstring(requests.get(path).text)\n\n # Find the title from the MEI file and update the Music21 Score metadata\n title = mei_doc.find('mei:meiHead//mei:titleStmt/mei:title', namespaces={\"mei\": MEINSURI}).text\n print(path, title)\n corpus.scores[i].metadata.title = title\n except:\n continue\n\nfor s in corpus.scores:\n print(s.metadata.title)",
"Requesting file from https://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_1.mei...\nSuccessfully imported.\nRequesting file from https://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_2.mei...\nSuccessfully imported.\nRequesting file from https://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_3.mei...\nSuccessfully imported.\nRequesting file from https://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_4.mei...\nSuccessfully imported.\nRequesting file from https://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_5.mei...\nSuccessfully imported.\nRequesting file from https://crimproject.org/mei/MEI_4.0/CRIM_Model_0001.mei...\nSuccessfully imported.\nhttps://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_1.mei Missa Vidi speciosam: Kyrie\nhttps://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_2.mei Missa Vidi speciosam: Gloria\nhttps://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_3.mei Missa Vidi speciosam: Credo\nhttps://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_4.mei Missa Vidi speciosam: Sanctus\nhttps://crimproject.org/mei/MEI_4.0/CRIM_Mass_0002_5.mei Missa Vidi speciosam: Agnus Dei\nhttps://crimproject.org/mei/MEI_4.0/CRIM_Model_0001.mei Veni speciosam\nMissa Vidi speciosam: Kyrie\nMissa Vidi speciosam: Gloria\nMissa Vidi speciosam: Credo\nMissa Vidi speciosam: Sanctus\nMissa Vidi speciosam: Agnus Dei\nVeni speciosam\n"
]
],
[
[
"\n## Select Actual or Incremental Durations\n\n#### About Rhythmic Durations\n\n- For `find_close_matches` and `find_exact_matches`, rhythmic variation/duration is displayed, but **not** factored into the calculation of matching.\n- **Incremental Offset** calculates the intervals using a **fixed offset between notes**, no matter their actual duration. Use this to ignore passing tones or other ornaments. The offsets are expressed in multiples of the quarter note (Offset = 1 samples at quarter note; Offset = 2 at half note, etc). Set with `vectors = IntervalBase(corpus.note_list_incremental_offset(2))`",
"_____no_output_____"
]
],
[
[
"vectors = IntervalBase(corpus.note_list)\n#vectors = IntervalBase(corpus.note_list_incremental_offset(2))",
"_____no_output_____"
]
],
[
[
"***\n\n## Select Generic or Semitone Scale:\n\n- **Length of the Soggetto**: `into_patterns([vectors.semitone_intervals], 5)` \n\n- The **number** in this command represents the **minimum number of vectors to find**. 5 vectors is 6 notes.\n",
"_____no_output_____"
]
],
[
[
"patterns = into_patterns([vectors.generic_intervals], 8)\n#patterns = into_patterns([vectors.semitone_intervals], 4)",
"_____no_output_____"
]
],
[
[
"***\n\n## Select Exact Matches Here, or Close Below\n#### (Use comment feature to select screen preview or CSV output) \n\n- **Exact** is exact in *all* ways `find_exact_matches(patterns, 2)` \n- The **number** in this command represents the **minimum number of matching melodies needed before reporting**. This allows us to filter for common or uncommon soggetti.",
"_____no_output_____"
]
],
[
[
"exact_matches = find_exact_matches(patterns, 3)\n# Use this for exact screen preview\n#for item in exact_matches:\n #item.print_exact_matches()\n\noutput_exact = export_pandas(exact_matches)\npd.DataFrame(output_exact).head()\noutput_exact[\"pattern_generating_match\"] = output_exact[\"pattern_generating_match\"].apply(tuple)\n\nresults = pd.DataFrame(output_exact)\nresults[\"pattern_generating_match\"] = results[\"pattern_generating_match\"].apply(tuple)\nresults\n#export_to_csv(exact_matches)",
"Finding exact matches...\n57 melodic intervals had more than 3 exact matches.\n\n"
]
],
[
[
"### A Quick Overview of the Results",
"_____no_output_____"
]
],
[
[
"total_matches = len(output_exact)\nunique_sogetti = output_exact.pattern_generating_match.apply(str).nunique()\nsummary = 'There are {} unique soggetti and {} total matches in this search'.format(unique_sogetti, total_matches)\nsummary",
"_____no_output_____"
]
],
[
[
"### Group by the Pattern Generating Match and Check Distribution of Results\n\n- Report Top Ten and Bottom Ten Results)",
"_____no_output_____"
]
],
[
[
"pattern_inventory = pd.DataFrame(output_exact.groupby(\"pattern_generating_match\").size().sort_values(ascending=False)[:10])\npattern_inventory",
"_____no_output_____"
],
[
"pattern_inventory = pd.DataFrame(output_exact.groupby(\"pattern_generating_match\").size().sort_values(ascending=True)[:10])\npattern_inventory",
"_____no_output_____"
]
],
[
[
"***\n\n### Select Close Matches Here\n#### (Comment out the 'for item iteration' in order to skip screen preview)\n\n- **Close** matches allow for melodic variation (see more below). `find_close_matches(patterns, 2, 1)`\n- The **first number** in this command is the **minimum number of melodies** needed before reporting\n- The **second number** is **threshold of similarity** needed in order to find a match. \n- Lower number = very similar; higher number = less similar\n\n##### More about Close Matches \n- The **threshold for close matches** is determined by the **second number** called in the method. \n- We select two patterns, then compare *each vector in each pattern successively*. \n- The *differences between each vector are summed*. \n- If that value is **below the threshold specified**, we consider the **two patterns closely matched**.\n- The format of the method call is `find_close_matches(the array you get from into_patterns, minimum matches needed to be displayed, threshold for close match)`.",
"_____no_output_____"
]
],
[
[
"close_matches = find_close_matches(patterns, 2, 1)\n#for item in close_matches:\n #item.print_close_matches()\n #return pd.DataFrame(close_matches)\n\noutput_close = export_pandas(close_matches)\noutput_close[\"pattern_generating_match\"] = output_close[\"pattern_generating_match\"].apply(tuple)\n\nresults = pd.DataFrame(output_close)\nresults[\"pattern_generating_match\"] = results[\"pattern_generating_match\"].apply(tuple)\nresults.head(50)\n#export_to_csv(close_matches)",
"Finding close matches...\n644 melodic intervals had more than 2 exact or close matches.\n\n"
]
],
[
[
"### How Many Unique Soggetti? How many instances?",
"_____no_output_____"
]
],
[
[
"total_matches = len(output_close)\nunique_sogetti = output_close.pattern_generating_match.apply(str).nunique()\nsummary = 'There are {} unique soggetti and {} total matches in this search'.format(unique_sogetti, total_matches)\nsummary",
"_____no_output_____"
]
],
[
[
"### Top and Bottom Ten Soggetti\n\n\n",
"_____no_output_____"
]
],
[
[
"pattern_inventory = pd.DataFrame(output_close.groupby(\"pattern_generating_match\").size().sort_values(ascending=False)[:50])\npattern_inventory",
"_____no_output_____"
],
[
"pattern_inventory = pd.DataFrame(output_close.groupby(\"pattern_generating_match\").size().sort_values(ascending=True)[:10])\npattern_inventory\n",
"_____no_output_____"
]
],
[
[
"***\n\n### Classify Patterns Here \n#### Note: depends on choice of Close or Exact above! Must choose appropriate one below!\n#### Enable \"export_to_csv\" line to allow this within Notebook (must answer \"Y\" and provide filename)",
"_____no_output_____"
]
],
[
[
"%%capture\nclassify_matches(close_matches, 2)\n#classify_matches(exact_matches, 2)\ncm = classify_matches(close_matches, 2)\n#pd.DataFrame(classified_matches)\noutput_cm = export_pandas(cm)\n#pd.DataFrame(output).head()\n\n## For CSV export, use the following (and follow prompts for file name)\n\n#export_to_csv(cm)\n\n",
"_____no_output_____"
],
[
"short_out = output_cm.drop(columns=[\"ema_url\"])\nshort_out",
"_____no_output_____"
],
[
"def classified_matches_to_pandas(matches):\n \n soggetti_matches = []\n \n for i, cm in enumerate(matches):\n \n for j, soggetti in enumerate(cm.matches):\n \n soggetti_matches.append({\n \"piece\": soggetti.first_note.metadata.title,\n \"type\": cm.type,\n \"part\": soggetti.first_note.part.strip(\"[] \"),\n \"bar\": soggetti.first_note.note.measureNumber,\n \"entry_number\": j + 1,\n \"pattern\": cm.pattern,\n \"match_number\": i + 1\n })\n return pd.DataFrame(soggetti_matches)",
"_____no_output_____"
],
[
"df = classified_matches_to_pandas(cm)\npd.set_option('display.max_rows', 50)\ndf.head(10)",
"_____no_output_____"
],
[
"wide_df = df.pivot_table(index=[\"match_number\", \"piece\", \"type\"],\n columns=\"entry_number\",\n values=[\"part\", \"bar\"],\n aggfunc=lambda x: x)\n\nwide_df.columns = [f\"{a}_{b}\" for a, b in wide_df.columns]\n\nwide_df.head().reset_index()\n\nwide_df.shape\n\nwide_df",
"_____no_output_____"
]
],
[
[
"## Read CSV of Classified Matches\n\n- Update file name to match the output of previous cells for Classifier",
"_____no_output_____"
]
],
[
[
"results = pd.read_csv('Sandrin_Classified.csv')\nresults.rename(columns=\n {'Pattern Generating Match': 'Pattern_Generating_Match', \n 'Pattern matched':'Pattern_Matched',\n 'Classification Type': 'Classification_Type',\n 'Piece Title': 'Piece_Title',\n 'First Note Measure Number': 'Start_Measure',\n 'Last Note Measure Number': 'Stop_Measure',\n 'Note Durations': 'Note_Durations'\n },\n inplace=True)\n\nresults['note_durations'] = results['note_durations'].apply(ast.literal_eval)\n\ndurations = results['note_durations']\nresults.head()",
"_____no_output_____"
]
],
[
[
"# Durational Ratios\n\n#### This Function Calculates the Ratios of the Durations in each Match",
"_____no_output_____"
]
],
[
[
"# makes pairs of ratio strings\n\ndef pairwise(iterable):\n \"s -> (s0,s1), (s1,s2), (s2, s3), ...\"\n a, b = tee(iterable)\n next(b, None)\n return zip(a, b)\n\ndef get_ratios(input_list):\n ratio_pairs = []\n for a, b in pairwise(input_list):\n ratio_pairs.append(b / a)\n return ratio_pairs\n\n\n",
"_____no_output_____"
]
],
[
[
"#### Now call the function to operate on the RESULTS file from earlier",
"_____no_output_____"
]
],
[
[
"# calculates 'duration ratios' for each soggetto, then adds this to the DF\n\nresults[\"duration_ratios\"] = results.note_durations.apply(get_ratios)\nshort_results = results.drop(columns=[\"ema_url\"])\nshort_out.head(10)",
"_____no_output_____"
]
],
[
[
"## Group by the Pattern Generating Match\n- Each has its own string of durations, and duration ratios\n- and then we compare the ratios to get the differences\n- the \"list(combinations)\" method takes care of building the pairs, using data from our dataframe 'results'",
"_____no_output_____"
]
],
[
[
"def compare_ratios(ratios_1, ratios_2):\n \n ## division of lists \n # using zip() + list comprehension \n diffs = [i - j for i, j in zip(ratios_1, ratios_2)] \n abs_diffs = [abs(ele) for ele in diffs] \n sum_diffs = sum(abs_diffs)\n\n return sum_diffs\n\n#results[\"Pattern_Generating_Match\"] = results[\"Pattern_Generating_Match\"].apply(tuple) \n\ndef get_ratio_distances(results, pattern_col, output_cols):\n \n matches = []\n\n for name, group in results.groupby(pattern_col):\n\n ratio_pairs = list(combinations(group.index.values, 2))\n\n for a, b in ratio_pairs:\n \n a_match = results.loc[a]\n b_match = results.loc[b]\n \n sum_diffs = compare_ratios(a_match.duration_ratios, b_match.duration_ratios)\n \n match_dict = {\n \"pattern\": name,\n \"sum_diffs\": sum_diffs\n }\n \n for col in output_cols:\n match_dict.update({\n f\"match_1_{col}\": a_match[col],\n f\"match_2_{col}\": b_match[col]\n })\n \n matches.append(match_dict)\n \n return pd.DataFrame(matches)",
"_____no_output_____"
]
],
[
[
"### Now Run the Function to get the 'edit distances' for the durations of matching patterns",
"_____no_output_____"
]
],
[
[
"ratio_distances = get_ratio_distances(results, \"pattern_generating_match\", [\"piece_title\", \"part\", \"start_measure\", \"end_measure\"])\nratio_distances.head()",
"_____no_output_____"
]
],
[
[
"### And FILTER the results according to any threshold we like",
"_____no_output_____"
]
],
[
[
"ratios_filtered = ratio_distances[ratio_distances.sum_diffs <= 1]\nratios_filtered",
"_____no_output_____"
]
],
[
[
"### Now Group the Duration-Filter Results by the Pattern (which shows us very closely related soggetti in sets)",
"_____no_output_____"
]
],
[
[
"grouped = ratios_filtered.groupby(\"pattern\")\ngrouped.head()",
"_____no_output_____"
],
[
"ratios_filtered.to_csv(\"filtered_sample_pair.csv\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
d0b7d6fee9c50eb825e90bc7ce93140b3a7fa3a6 | 6,138 | ipynb | Jupyter Notebook | lessons/ObjectOrientedProgramming/JupyterNotebooks/1.OOP_syntax_shirt_practice/shirt_exercise.ipynb | thomasdunlap/DSND_Term2 | 953e1eef1e7d9568bd02b18a8a16a034a0a3f369 | [
"MIT"
] | null | null | null | lessons/ObjectOrientedProgramming/JupyterNotebooks/1.OOP_syntax_shirt_practice/shirt_exercise.ipynb | thomasdunlap/DSND_Term2 | 953e1eef1e7d9568bd02b18a8a16a034a0a3f369 | [
"MIT"
] | 1 | 2021-06-02T00:33:12.000Z | 2021-06-02T00:33:12.000Z | lessons/ObjectOrientedProgramming/JupyterNotebooks/1.OOP_syntax_shirt_practice/shirt_exercise.ipynb | thomasdunlap/DSND_Term2 | 953e1eef1e7d9568bd02b18a8a16a034a0a3f369 | [
"MIT"
] | 1 | 2020-06-02T01:13:26.000Z | 2020-06-02T01:13:26.000Z | 30.69 | 289 | 0.57869 | [
[
[
"# Use the Shirt Class\n\nYou've seen what a class looks like and how to instantiate an object. Now it's your turn to write code that insantiates a shirt object.\n\n# Explanation of the Code\nThis Jupyter notebook is inside of a folder called 1.OOP_syntax_shirt_practice. You can see the folder if you click on the \"Jupyter\" logo above the notebook. Inside the folder are three files:\n- shirt_exercise.ipynb, which is the file you are currently looking at\n- answer.py containing answers to the exercise\n- tests.py, tests for checking your code - you can run these tests using the last code cell at the bottom of this notebook\n\n# Your Task\nThe shirt_exercise.ipynb file, which you are currently looking at if you are reading this, has an exercise to help guide you through coding with an object in Python.\n\nFill out the TODOs in each section of the Jupyter notebook. You can find a solution in the answer.py file.\n\nFirst, run this code cell below to load the Shirt class.",
"_____no_output_____"
]
],
[
[
"class Shirt:\n\n def __init__(self, shirt_color, shirt_size, shirt_style, shirt_price):\n self.color = shirt_color\n self.size = shirt_size\n self.style = shirt_style\n self.price = shirt_price\n \n def change_price(self, new_price):\n \n self.price = new_price\n \n def discount(self, discount):\n\n return self.price * (1 - discount)",
"_____no_output_____"
],
[
"### TODO:\n# - insantiate a shirt object with the following characteristics:\n# - color red, size S, style long-sleeve, and price 25\n# - store the object in a variable called shirt_one\n#\n#\n###\nshirt_one = Shirt(shirt_color='red', shirt_size= 'S', shirt_style='long-sleeve', shirt_price=25)",
"_____no_output_____"
],
[
"### TODO:\n# - print the price of the shirt using the price attribute\n# - use the change_price method to change the price of the shirt to 10\n# - print the price of the shirt using the price attribute\n# - use the discount method to print the price of the shirt with a 12% discount\n#\n###\nprint(shirt_one.price)\nshirt_one.change_price(10)\nprint(shirt_one.price)\nprint(shirt_one.discount(12))",
"25\n10\n-110\n"
],
[
"### TODO:\n#\n# - instantiate another object with the following characteristics:\n# . - color orange, size L, style short-sleeve, and price 10\n# - store the object in a variable called shirt_two\n#\n###\nshirt_two = Shirt('orange', 'L', 'short-sleeve', 10)",
"_____no_output_____"
],
[
"### TODO:\n#\n# - calculate the total cost of shirt_one and shirt_two\n# - store the results in a variable called total\n# \n###\ntotal = shirt_two.price + shirt_one.price",
"_____no_output_____"
],
[
"### TODO:\n#\n# - use the shirt discount method to calculate the total cost if\n# shirt_one has a discount of 14% and shirt_two has a discount\n# of 6%\n# - store the results in a variable called total_discount\n###\ntotal_discount = shirt_one.discount(.14) + shirt_two.discount(.06)",
"_____no_output_____"
]
],
[
[
"# Test your Code\n\n\nThe following code cell tests your code. \n\nThere is a file called tests.py containing a function called run_tests(). The run_tests() function executes a handful of assert statements to check your work. You can see this file if you go to the Jupyter Notebook menu and click on \"File->Open\" and then open the tests.py file.\n\nExecute the next code cell. The code will produce an error if your answers in this exercise are not what was expected. Keep working on your code until all tests are passing.\n\nIf you run the code cell and there is no output, then you passed all the tests!\n\nAs mentioned previously, there's also a file with a solution. To find the solution, click on the Jupyter logo at the top of the workspace, and then enter the folder titled 1.OOP_syntax_shirt_practice",
"_____no_output_____"
]
],
[
[
"# Unit tests to check your solution\nfrom tests import run_tests\n\nrun_tests(shirt_one, shirt_two, total, total_discount)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
d0b7dadf996c7e572055ac7f36cd1e1e04a265dd | 8,497 | ipynb | Jupyter Notebook | Complete-Python-3-Bootcamp-master/12-Advanced Python Modules/04-Python Debugger (pdb).ipynb | davidMartinVergues/PYTHON | dd39d3aabfc43b3cb09aadb2919e51d03364117d | [
"DOC"
] | 8 | 2020-09-02T03:59:02.000Z | 2022-01-08T23:36:19.000Z | Complete-Python-3-Bootcamp-master/12-Advanced Python Modules/04-Python Debugger (pdb).ipynb | davidMartinVergues/PYTHON | dd39d3aabfc43b3cb09aadb2919e51d03364117d | [
"DOC"
] | null | null | null | Complete-Python-3-Bootcamp-master/12-Advanced Python Modules/04-Python Debugger (pdb).ipynb | davidMartinVergues/PYTHON | dd39d3aabfc43b3cb09aadb2919e51d03364117d | [
"DOC"
] | 3 | 2020-11-18T12:13:05.000Z | 2021-02-24T19:31:50.000Z | 55.175325 | 1,417 | 0.623867 | [
[
[
"___\n\n<a href='https://www.udemy.com/user/joseportilla/'><img src='../Pierian_Data_Logo.png'/></a>\n___\n<center><em>Content Copyright by Pierian Data</em></center>",
"_____no_output_____"
],
[
"# Python Debugger\n\nYou've probably used a variety of print statements to try to find errors in your code. A better way of doing this is by using Python's built-in debugger module (pdb). The pdb module implements an interactive debugging environment for Python programs. It includes features to let you pause your program, look at the values of variables, and watch program execution step-by-step, so you can understand what your program actually does and find bugs in the logic.\n\nThis is a bit difficult to show since it requires creating an error on purpose, but hopefully this simple example illustrates the power of the pdb module. <br>*Note: Keep in mind it would be pretty unusual to use pdb in an Jupyter Notebook setting.*\n\n___\nHere we will create an error on purpose, trying to add a list to an integer",
"_____no_output_____"
]
],
[
[
"x = [1,3,4]\ny = 2\nz = 3\n\nresult = y + z\nprint(result)\nresult2 = y+x\nprint(result2)",
"5\n"
]
],
[
[
"Hmmm, looks like we get an error! Let's implement a set_trace() using the pdb module. This will allow us to basically pause the code at the point of the trace and check if anything is wrong.",
"_____no_output_____"
]
],
[
[
"import pdb\n\nx = [1,3,4]\ny = 2\nz = 3\n\nresult = y + z\nprint(result)\n\n# Set a trace using Python Debugger\npdb.set_trace()\n\nresult2 = y+x\nprint(result2)",
"5\n--Return--\n> <ipython-input-2-1084246755fa>(11)<module>()->None\n-> pdb.set_trace()\n(Pdb) x\n[1, 3, 4]\n(Pdb) y\n2\n(Pdb) result2\n*** NameError: name 'result2' is not defined\n(Pdb) q\n"
]
],
[
[
"Great! Now we could check what the various variables were and check for errors. You can use 'q' to quit the debugger. For more information on general debugging techniques and more methods, check out the official documentation:\nhttps://docs.python.org/3/library/pdb.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b7ecb7b602c3116b961d04efb3cec4eba04e2f | 22,993 | ipynb | Jupyter Notebook | tutorials/05_scikit_learn.ipynb | sylvainlugeon/ntds_2019 | d16314bda9d0744980ef95e879f65cd4fb51b461 | [
"MIT"
] | 2 | 2020-01-15T20:36:55.000Z | 2020-04-19T09:06:46.000Z | tutorials/05_scikit_learn.ipynb | sylvainlugeon/ntds_2019 | d16314bda9d0744980ef95e879f65cd4fb51b461 | [
"MIT"
] | 1 | 2019-10-29T07:16:28.000Z | 2019-10-29T07:16:28.000Z | tutorials/05_scikit_learn.ipynb | sylvainlugeon/ntds_2019 | d16314bda9d0744980ef95e879f65cd4fb51b461 | [
"MIT"
] | 1 | 2019-10-16T01:17:28.000Z | 2019-10-16T01:17:28.000Z | 32.430183 | 417 | 0.595834 | [
[
[
"# [NTDS'19] tutorial 5: machine learning with scikit-learn\n[ntds'19]: https://github.com/mdeff/ntds_2019\n\n[Nicolas Aspert](https://people.epfl.ch/nicolas.aspert), [EPFL LTS2](https://lts2.epfl.ch).\n\n* Dataset: [digits](https://archive.ics.uci.edu/ml/datasets/Pen-Based+Recognition+of+Handwritten+Digits)\n* Tools: [scikit-learn](https://scikit-learn.org/stable/), [numpy](http://www.numpy.org), [scipy](https://www.scipy.org), [matplotlib](https://matplotlib.org)",
"_____no_output_____"
],
[
"*scikit-learn* is a machine learning python library. Most commonly used algorithms for classification, clustering and regression are implemented as part of the library, e.g.\n* [Logistic regression](https://en.wikipedia.org/wiki/Logistic_regression)\n* [k-means clustering](https://en.wikipedia.org/wiki/K-means_clustering)\n* [Support vector machines](https://en.wikipedia.org/wiki/Support-vector_machine)\n* ...\n\nThe aim of this tutorial is to show basic usage of some simple machine learning techniques. \nCheck the official [documentation](https://scikit-learn.org/stable/documentation.html) for more information, especially the [tutorials](https://scikit-learn.org/stable/tutorial/index.html) section.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport sklearn",
"_____no_output_____"
]
],
[
[
"## Data loading\n\nWe will use a dataset named *digits*.\nIt is made of 1797 handwritten digits images (of size 8x8 pixels each) acquired from 44 different writers. \nEach image is labelled according to the digit present in the image.\n\nYou can find more information about this dataset [here](https://archive.ics.uci.edu/ml/datasets/Pen-Based+Recognition+of+Handwritten+Digits).\n\n",
"_____no_output_____"
],
[
"Load the dataset.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_digits\n\ndigits = load_digits()",
"_____no_output_____"
]
],
[
[
"The `digits` variable contains several fields.\n\nIn `images` you have all samples as 2-dimensional arrays.",
"_____no_output_____"
]
],
[
[
"print(digits.images.shape)\nprint(digits.images[0])\nplt.imshow(digits.images[0], cmap=plt.cm.gray);",
"_____no_output_____"
]
],
[
[
"In `data`, the same samples are represented as 1-d vectors of length 64.",
"_____no_output_____"
]
],
[
[
"print(digits.data.shape)\nprint(digits.data[0])",
"_____no_output_____"
]
],
[
[
" In `target` you have the label corresponding to each image.",
"_____no_output_____"
]
],
[
[
"print(digits.target.shape)\nprint(digits.target)",
"_____no_output_____"
]
],
[
[
"Let us visualize the 20 first entries of the dataset (image display kept small on purpose)",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(15, 0.5))\nfor index, (image, label) in enumerate(zip(digits.images[0:20], digits.target[0:20])):\n ax = fig.add_subplot(1, 20, index+1)\n ax.imshow(image, cmap=plt.cm.gray)\n ax.set_title(label)\n ax.axis('off')",
"_____no_output_____"
]
],
[
[
"### Training/Test set\n\nBefore training our model, the [`train_test_split`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html) function will separate our dataset into a training set and a test set. The samples from the test set are never used during the training phase. This allows for a fair evaluation of the model's performance.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\ntrain_img, test_img, train_lbl, test_lbl = train_test_split(\n digits.data, digits.target, test_size=1/6) # keep ~300 images as test set",
"_____no_output_____"
]
],
[
[
"We can check that all classes are well balanced in the training and test sets.",
"_____no_output_____"
]
],
[
[
"np.histogram(train_lbl, bins=10)",
"_____no_output_____"
],
[
"np.histogram(test_lbl, bins=10)",
"_____no_output_____"
]
],
[
[
"## Supervised learning: logistic regression\n\n### Linear regression reminder\n\nLinear regression is used to predict an dependent value $y$ from an n-dimensional vector $x$.\nThe assumption made here is that the output depends linearly on the input components, i.e. $y = mx + b$.\n\nGiven a set of input and output values, the goal is to compute $m$ and $b$ minimizing the [mean squared error (MSE)](https://en.wikipedia.org/wiki/Mean_squared_error) between the predicted and actual outputs.\nIn scikit-learn this method is available through [`LinearRegression`](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html).\n\n### Logistic regression\n\nLogistic regression is used to predict categorical data (e.g. yes/no, member/non-member, ham/spam, benign/malignant, ...).\nIt uses the output of a linear predictor, and maps it to a probability using a [sigmoid function](https://en.wikipedia.org/wiki/Sigmoid_function), such as the logistic function $s(z) = \\frac{1}{1+e^{-z}}$. \nThe output is a probability score between 0 and 1, and using a simple thresholding the class output will be positive if the probability is greater than 0.5, negative if not.\nA [log-loss cost function](http://wiki.fast.ai/index.php/Logistic_Regression#Cost_Function) (not just the MSE as for linear regression) is used to train logistic regression (using gradient descent for instance).\n\n[Multinomial logistic regression](https://en.wikipedia.org/wiki/Multinomial_logistic_regression) is an extension of the binary classification problem to a $n$-classes problem.",
"_____no_output_____"
],
[
"We can now create a logistic regression object and fit the parameters using the training data.\n\nNB: as the dataset is quite simple, default parameters will give good results. Check the [documentation](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html) for fine-tuning possibilities.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\n# All unspecified parameters are left to their default values.\nlogisticRegr = LogisticRegression(verbose=1, solver='liblinear', multi_class='auto') # set solver and multi_class to silence warnings",
"_____no_output_____"
],
[
"logisticRegr.fit(train_img, train_lbl)",
"_____no_output_____"
]
],
[
[
"## Model performance evaluation\n\nFor a binary classification problem, let us denote by $TP$, $TN$, $FP$, and $FN$ the number of true positives, true negatives, false positives and false negatives.\n\n### Accuracy\n\nThe *accuracy* is defined by $a = \\frac{TP}{TP + TN + FP + FN}$\n\nNB: in scikit-learn, models may have different definitions of the `score` method. For multi-class logistic regression, the value is the mean accuracy for each class.",
"_____no_output_____"
]
],
[
[
"score = logisticRegr.score(test_img, test_lbl)\nprint(f'accuracy = {score:.4f}')",
"_____no_output_____"
]
],
[
[
"### F1 score\n\nAccuracy only provides partial information about the performance of a model. Many other [metrics](https://scikit-learn.org/stable/modules/model_evaluation.html#classification-metrics) are part of scikit-learn.\n\nA metric that provides a more complete overview of the classification performance is the [F1 score](https://en.wikipedia.org/wiki/F1_score). It takes into account not only the valid predictions but also the incorrect ones, by combining precision and recall.\n\n*Precision* is the number of positive predictions divided by the total number of positive class values predicted, i.e. $p=\\frac{TP}{TP+FP}$. A low precision indicates a high number of false positives.\n\n*Recall* is the number of positive predictions divided by the number of positive class values in the test data, i.e. $r=\\frac{TP}{TP+FN}$. A low recall indicates a high number of false negatives.\n\nFinally the F1 score is the harmonic mean between precision and recall, i.e. $F1=2\\frac{p.r}{p+r}$",
"_____no_output_____"
],
[
"Let us compute the predicted labels in the test set:",
"_____no_output_____"
]
],
[
[
"pred_lbl = logisticRegr.predict(test_img)",
"_____no_output_____"
],
[
"from sklearn.metrics import f1_score, classification_report\nfrom sklearn.utils.multiclass import unique_labels",
"_____no_output_____"
]
],
[
[
"The [`f1_score`](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html#sklearn.metrics.f1_score) function computes the F1 score. The `average` parameter controls whether the result is computed globally over all classes (`average='micro'`) or if the F1 score is computed for each class then averaged (`average='macro'`).",
"_____no_output_____"
]
],
[
[
"f1_score(test_lbl, pred_lbl, average='micro')",
"_____no_output_____"
],
[
"f1_score(test_lbl, pred_lbl, average='macro')",
"_____no_output_____"
]
],
[
[
"`classification_report` provides a synthetic overview of all results for each class, as well as globally.",
"_____no_output_____"
]
],
[
[
"print(classification_report(test_lbl, pred_lbl))",
"_____no_output_____"
]
],
[
[
"### Confusion matrix\n\nIn the case of a multi-class problem, the *confusion matrix* is often used to present the results.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix\n\ndef plot_confusion_matrix(y_true, y_pred, classes,\n normalize=False,\n title=None,\n cmap=plt.cm.Blues):\n \"\"\"\n This function prints and plots the confusion matrix.\n Normalization can be applied by setting `normalize=True`.\n \"\"\"\n if not title:\n if normalize:\n title = 'Normalized confusion matrix'\n else:\n title = 'Confusion matrix, without normalization'\n\n # Compute confusion matrix\n cm = confusion_matrix(y_true, y_pred)\n # Only use the labels that appear in the data\n classes = classes[unique_labels(y_true, y_pred)]\n if normalize:\n cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]\n print(\"Normalized confusion matrix\")\n else:\n print('Confusion matrix, without normalization')\n\n print(cm)\n\n fig, ax = plt.subplots()\n im = ax.imshow(cm, interpolation='nearest', cmap=cmap)\n ax.figure.colorbar(im, ax=ax)\n # We want to show all ticks...\n ax.set(xticks=np.arange(cm.shape[1]),\n yticks=np.arange(cm.shape[0]),\n # ... and label them with the respective list entries\n xticklabels=classes, yticklabels=classes,\n title=title,\n ylabel='True label',\n xlabel='Predicted label')\n\n # Rotate the tick labels and set their alignment.\n plt.setp(ax.get_xticklabels(), rotation=45, ha=\"right\",\n rotation_mode=\"anchor\")\n\n # Loop over data dimensions and create text annotations.\n fmt = '.2f' if normalize else 'd'\n thresh = cm.max() / 2.\n for i in range(cm.shape[0]):\n for j in range(cm.shape[1]):\n ax.text(j, i, format(cm[i, j], fmt),\n ha=\"center\", va=\"center\",\n color=\"white\" if cm[i, j] > thresh else \"black\")\n fig.tight_layout()\n return ax",
"_____no_output_____"
],
[
"plot_confusion_matrix(test_lbl, pred_lbl, np.array(list(map(lambda x: str(x), range(10)))), normalize=False)",
"_____no_output_____"
]
],
[
[
"## Supervised learning: support-vector machines\n\n[Support-vector machines (SVM)](https://en.wikipedia.org/wiki/Support-vector_machine) are also used for classification tasks.\nFor a binary classification task of $n$-dimensional feature vectors, a linear SVM try to return the ($n-1$)-dimensional hyperplane that separate the two classes with the largest possible margin.\nNonlinear SVMs fit the maximum-margin hyperplane in a transformed feature space.\nAlthough the classifier is a hyperplane in the transformed feature space, it may be nonlinear in the original input space. \n\nThe goal here is to show that a method (e.g. the previously used logistic regression) can be substituted transparently for another one.",
"_____no_output_____"
]
],
[
[
"from sklearn import svm",
"_____no_output_____"
]
],
[
[
"Default parameters perform well on this dataset.\nIt might be needed to adjust $C$ and $\\gamma$ (e.g. via a grid search) for optimal performance (cf. [SVC documentation](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html#sklearn.svm.SVC)).",
"_____no_output_____"
]
],
[
[
"clf = svm.SVC(gamma='scale') # default kernel is RBF",
"_____no_output_____"
],
[
"clf.fit(train_img, train_lbl)",
"_____no_output_____"
]
],
[
[
"The classification accuracy improves with respect to logistic regression (here `score` also computes mean accuracy, as in logistic regression).",
"_____no_output_____"
]
],
[
[
"clf.score(test_img, test_lbl)",
"_____no_output_____"
]
],
[
[
"The F1 score is also improved.",
"_____no_output_____"
]
],
[
[
"pred_lbl_svm = clf.predict(test_img)\nprint(classification_report(test_lbl, pred_lbl_svm))",
"_____no_output_____"
]
],
[
[
"## Unsupervised learning: $k$-means\n\n[$k$-means](https://en.wikipedia.org/wiki/K-means_clustering) aims at partitioning a samples into $k$ clusters, s.t. each sample belongs to the cluster having the closest mean. Its implementation is iterative, and relies on a prior knowledge of the number of clusters present. \n\nOne important step in $k$-means clustering is the initialization, i.e. the choice of initial clusters to be refined.\nThis choice can have a significant impact on results.",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans",
"_____no_output_____"
],
[
"kmeans = KMeans(n_clusters=10)",
"_____no_output_____"
],
[
"kmeans.fit(digits.data)\nkm_labels = kmeans.predict(digits.data)",
"_____no_output_____"
],
[
"digits.target",
"_____no_output_____"
],
[
"km_labels",
"_____no_output_____"
]
],
[
[
"Since we have ground truth information of classes, we can check if the $k$-means results make sense.\nHowever as you can see, the labels produced by $k$-means and the ground truth ones do not match.\nAn agreement score based on [mutual information](https://scikit-learn.org/stable/modules/clustering.html#clustering-evaluation), insensitive to labels permutation can be used to evaluate the results. ",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import adjusted_mutual_info_score",
"_____no_output_____"
],
[
"adjusted_mutual_info_score(digits.target, kmeans.labels_)",
"_____no_output_____"
]
],
[
[
"## Unsupervized learning: dimensionality reduction\n\nYou can also try to visualize the clusters as in this [scikit-learn demo](https://scikit-learn.org/stable/auto_examples/cluster/plot_kmeans_digits.html). Mapping the input features to lower dimensional embeddings (2D or 3D), e.g. using PCA otr tSNE is required for visualization. [This demo](https://scikit-learn.org/stable/auto_examples/manifold/plot_lle_digits.html) provides an overview of the possibilities.",
"_____no_output_____"
]
],
[
[
"from matplotlib import offsetbox\n\ndef plot_embedding(X, y, title=None):\n \"\"\"Scale and visualize the embedding vectors.\"\"\"\n x_min, x_max = np.min(X, 0), np.max(X, 0)\n X = (X - x_min) / (x_max - x_min)\n\n plt.figure()\n ax = plt.subplot(111)\n for i in range(X.shape[0]):\n plt.text(X[i, 0], X[i, 1], str(y[i]),\n color=plt.cm.Set1(y[i] / 10.),\n fontdict={'weight': 'bold', 'size': 9})\n\n if hasattr(offsetbox, 'AnnotationBbox'):\n # only print thumbnails with matplotlib > 1.0\n shown_images = np.array([[1., 1.]]) # just something big\n for i in range(X.shape[0]):\n dist = np.sum((X[i] - shown_images) ** 2, 1)\n if np.min(dist) < 4e-3:\n # don't show points that are too close\n continue\n shown_images = np.r_[shown_images, [X[i]]]\n imagebox = offsetbox.AnnotationBbox(\n offsetbox.OffsetImage(digits.images[i], cmap=plt.cm.gray_r),\n X[i])\n ax.add_artist(imagebox)\n plt.xticks([]), plt.yticks([])\n if title is not None:\n plt.title(title)",
"_____no_output_____"
],
[
"from sklearn import manifold",
"_____no_output_____"
],
[
"tsne = manifold.TSNE(n_components=2, init='pca', random_state=0)\nX_tsne = tsne.fit_transform(digits.data)\n\nplot_embedding(X_tsne, digits.target,\n \"t-SNE embedding of the digits (ground truth labels)\")",
"_____no_output_____"
],
[
"plot_embedding(X_tsne, km_labels,\n \"t-SNE embedding of the digits (kmeans labels)\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
d0b803130db3c5f576841f1f22251d6bd94a8e45 | 7,322 | ipynb | Jupyter Notebook | stable/_downloads/d327bd04605fdc3280dac732219b5db7/plot_artifacts_detection.ipynb | drammock/mne-tools.github.io | 5d3a104d174255644d8d5335f58036e32695e85d | [
"BSD-3-Clause"
] | null | null | null | stable/_downloads/d327bd04605fdc3280dac732219b5db7/plot_artifacts_detection.ipynb | drammock/mne-tools.github.io | 5d3a104d174255644d8d5335f58036e32695e85d | [
"BSD-3-Clause"
] | null | null | null | stable/_downloads/d327bd04605fdc3280dac732219b5db7/plot_artifacts_detection.ipynb | drammock/mne-tools.github.io | 5d3a104d174255644d8d5335f58036e32695e85d | [
"BSD-3-Clause"
] | null | null | null | 49.809524 | 2,788 | 0.641765 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n\nIntroduction to artifacts and artifact detection\n================================================\n\nSince MNE supports the data of many different acquisition systems, the\nparticular artifacts in your data might behave very differently from the\nartifacts you can observe in our tutorials and examples.\n\nTherefore you should be aware of the different approaches and of\nthe variability of artifact rejection (automatic/manual) procedures described\nonwards. At the end consider always to visually inspect your data\nafter artifact rejection or correction.\n\nBackground: what is an artifact?\n--------------------------------\n\nArtifacts are signal interference that can be\nendogenous (biological) and exogenous (environmental).\nTypical biological artifacts are head movements, eye blinks\nor eye movements, heart beats. The most common environmental\nartifact is due to the power line, the so-called *line noise*.\n\nHow to handle artifacts?\n------------------------\n\nMNE deals with artifacts by first identifying them, and subsequently removing\nthem. Detection of artifacts can be done visually, or using automatic routines\n(or a combination of both). After you know what the artifacts are, you need\nremove them. This can be done by:\n\n - *ignoring* the piece of corrupted data\n - *fixing* the corrupted data\n\nFor the artifact detection the functions MNE provides depend on whether\nyour data is continuous (Raw) or epoch-based (Epochs) and depending on\nwhether your data is stored on disk or already in memory.\n\nDetecting the artifacts without reading the complete data into memory allows\nyou to work with datasets that are too large to fit in memory all at once.\nDetecting the artifacts in continuous data allows you to apply filters\n(e.g. a band-pass filter to zoom in on the muscle artifacts on the temporal\nchannels) without having to worry about edge effects due to the filter\n(i.e. filter ringing). Having the data in memory after segmenting/epoching is\nhowever a very efficient way of browsing through the data which helps\nin visualizing. So to conclude, there is not a single most optimal manner\nto detect the artifacts: it just depends on the data properties and your\nown preferences.\n\nIn this tutorial we show how to detect artifacts visually and automatically.\nFor how to correct artifacts by rejection see `tut-artifact-rejection`.\nTo discover how to correct certain artifacts by filtering see\n`tut-filter-resample` and to learn how to correct artifacts\nwith subspace methods like SSP and ICA see\n`tut-artifact-ssp` and `tut-artifact-ica`.\n\n\nArtifacts Detection\n-------------------\n\nThis tutorial discusses a couple of major artifacts that most analyses\nhave to deal with and demonstrates how to detect them.\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nimport mne\nfrom mne.datasets import sample\nfrom mne.preprocessing import create_ecg_epochs, create_eog_epochs\n\n# getting some data ready\ndata_path = sample.data_path()\nraw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'\n\nraw = mne.io.read_raw_fif(raw_fname, preload=True)",
"_____no_output_____"
]
],
[
[
"Low frequency drifts and line noise\n\n",
"_____no_output_____"
]
],
[
[
"(raw.copy().pick_types(meg='mag')\n .del_proj(0)\n .plot(duration=60, n_channels=100, remove_dc=False))",
"_____no_output_____"
]
],
[
[
"we see high amplitude undulations in low frequencies, spanning across tens of\nseconds\n\n",
"_____no_output_____"
]
],
[
[
"raw.plot_psd(tmax=np.inf, fmax=250)",
"_____no_output_____"
]
],
[
[
"On MEG sensors we see narrow frequency peaks at 60, 120, 180, 240 Hz,\nrelated to line noise.\nBut also some high amplitude signals between 25 and 32 Hz, hinting at other\nbiological artifacts such as ECG. These can be most easily detected in the\ntime domain using MNE helper functions\n\nSee `tut-filter-resample`.\n\n",
"_____no_output_____"
],
[
"ECG\n---\n\nfinds ECG events, creates epochs, averages and plots\n\n",
"_____no_output_____"
]
],
[
[
"average_ecg = create_ecg_epochs(raw).average()\nprint('We found %i ECG events' % average_ecg.nave)\njoint_kwargs = dict(ts_args=dict(time_unit='s'),\n topomap_args=dict(time_unit='s'))\naverage_ecg.plot_joint(**joint_kwargs)",
"_____no_output_____"
]
],
[
[
"we can see typical time courses and non dipolar topographies\nnot the order of magnitude of the average artifact related signal and\ncompare this to what you observe for brain signals\n\n",
"_____no_output_____"
],
[
"EOG\n---\n\n",
"_____no_output_____"
]
],
[
[
"average_eog = create_eog_epochs(raw).average()\nprint('We found %i EOG events' % average_eog.nave)\naverage_eog.plot_joint(**joint_kwargs)",
"_____no_output_____"
]
],
[
[
"Knowing these artifact patterns is of paramount importance when\njudging about the quality of artifact removal techniques such as SSP or ICA.\nAs a rule of thumb you need artifact amplitudes orders of magnitude higher\nthan your signal of interest and you need a few of such events in order\nto find decompositions that allow you to estimate and remove patterns related\nto artifacts.\n\nConsider the following tutorials for correcting this class of artifacts:\n- `tut-filter-resample`\n- `tut-artifact-ica`\n- `tut-artifact-ssp`\n\n",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b805bda4cca6c927c0d562d95249c9cb634734 | 3,128 | ipynb | Jupyter Notebook | nb/env.ipynb | fuzzyklein/hw-4.2.0 | 76a7fcae40c60f18b067d0f51ee70d7a578ffea0 | [
"Apache-2.0"
] | null | null | null | nb/env.ipynb | fuzzyklein/hw-4.2.0 | 76a7fcae40c60f18b067d0f51ee70d7a578ffea0 | [
"Apache-2.0"
] | null | null | null | nb/env.ipynb | fuzzyklein/hw-4.2.0 | 76a7fcae40c60f18b067d0f51ee70d7a578ffea0 | [
"Apache-2.0"
] | null | null | null | 21.135135 | 81 | 0.503197 | [
[
[
"!echo $PYTHONSTARTUP",
"\n"
],
[
"from os import environ",
"_____no_output_____"
],
[
"'PYTHONSTARTUP' in environ.keys()",
"_____no_output_____"
],
[
"import sys\nsys.path",
"_____no_output_____"
],
[
"[s for s in environ.keys() if s.startswith('XDG')]",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
d0b8101bca9b595b4a01f0fbea39e7b19d9c0aed | 3,394 | ipynb | Jupyter Notebook | tutorials/Caffe2OnnxExport.ipynb | oblanchet/tutorials | febfbe7840309dba4aca711177565e777296a2a6 | [
"Apache-2.0"
] | 2,728 | 2017-11-17T01:19:46.000Z | 2022-03-31T19:23:32.000Z | tutorials/Caffe2OnnxExport.ipynb | oblanchet/tutorials | febfbe7840309dba4aca711177565e777296a2a6 | [
"Apache-2.0"
] | 234 | 2017-11-21T21:21:42.000Z | 2022-03-31T15:25:12.000Z | tutorials/Caffe2OnnxExport.ipynb | oblanchet/tutorials | febfbe7840309dba4aca711177565e777296a2a6 | [
"Apache-2.0"
] | 578 | 2017-11-18T17:22:05.000Z | 2022-03-29T08:57:03.000Z | 27.370968 | 317 | 0.571892 | [
[
[
"# Exporting models from Caffe2 to ONNX\n\nIn this tutorial we are going to show you how to export a Caffe2 model to ONNX. You can either\n\n- Convert a Caffe2 model to an ONNX model in Python\n\nor \n\n- Convert a Caffe2 model file to an ONNX model file in the shell\n\nWe are going to use the squeezenet model in Caffe2 model zoo, its model files can be downloaded by running:\n\n```shell\n$ python -m caffe2.python.models.download squeezenet\n```",
"_____no_output_____"
],
[
"### Installation\n\n`onnx-caffe2` is now integrated as part of `caffe2` under `caffe2/python/onnx`.",
"_____no_output_____"
],
[
"### Note\n\nIn ONNX, the type and shape of the inputs and outpus are required to be presented in the model, while in Caffe2, they are not stored in the model files. So when doing the convertion, we need to provide these extra information to onnx-caffe2 (through a dictionary in Python/a json string in the shell interface).",
"_____no_output_____"
],
[
"### Exporting in Python",
"_____no_output_____"
]
],
[
[
"import onnx\nimport caffe2.python.onnx.frontend\nfrom caffe2.proto import caffe2_pb2\n\n# We need to provide type and shape of the model inputs, \n# see above Note section for explanation\ndata_type = onnx.TensorProto.FLOAT\ndata_shape = (1, 3, 224, 224)\nvalue_info = {\n 'data': (data_type, data_shape)\n}\n\npredict_net = caffe2_pb2.NetDef()\nwith open('predict_net.pb', 'rb') as f:\n predict_net.ParseFromString(f.read())\n\ninit_net = caffe2_pb2.NetDef()\nwith open('init_net.pb', 'rb') as f:\n init_net.ParseFromString(f.read())\n\nonnx_model = caffe2.python.onnx.frontend.caffe2_net_to_onnx_model(\n predict_net,\n init_net,\n value_info,\n)\n\nonnx.checker.check_model(onnx_model)",
"_____no_output_____"
]
],
[
[
"### Exporting in shell\n\n`onnx-caffe2` has bundled a shell command `convert-caffe2-to-onnx` for exporting Caffe2 model file to ONNX model file.\n\n```shell\n\n$ convert-caffe2-to-onnx predict_net.pb --caffe2-init-net init_net.pb --value-info '{\"data\": [1, [1, 3, 224, 224]]}' -o sqeezenet.onnx\n\n```\n\nRegarding to the `--value-info` flag, see above Note section for explanation.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b815df52c8430ec59f24e93dd371c9f3ada2a2 | 318,197 | ipynb | Jupyter Notebook | notebooks/station_map.ipynb | kylermurphy/gmag | c0e10074d73758beeecdc0b72abafd297ce1af22 | [
"MIT"
] | 1 | 2020-05-13T19:56:54.000Z | 2020-05-13T19:56:54.000Z | notebooks/station_map.ipynb | kylermurphy/gmag | c0e10074d73758beeecdc0b72abafd297ce1af22 | [
"MIT"
] | null | null | null | notebooks/station_map.ipynb | kylermurphy/gmag | c0e10074d73758beeecdc0b72abafd297ce1af22 | [
"MIT"
] | 1 | 2020-09-30T16:01:46.000Z | 2020-09-30T16:01:46.000Z | 153.200289 | 276,515 | 0.930006 | [
[
[
"from IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\n\nimport pandas as pd\nimport folium",
"_____no_output_____"
],
[
"# read station data\ndata = pd.read_csv('..\\\\gmag\\\\Stations\\\\station_list.csv')\narrays = data.Array.unique()\narrays",
"_____no_output_____"
],
[
"stn_map = folium.Map(location=[40,200],zoom_start=1,\n width=650, height=400)\n\ncol = ['red','lightred','orange','cadetblue',\n 'blue','lightblue','purple','lightgreen','lightgray'\n 'grey','darkblue']\n\n\n\nfor x, y in zip(arrays,col): \n stn_dat = data[data['Array'] == x].copy()\n for label, row in stn_dat.iterrows():\n folium.Marker([row['Latitude'],row['Longitude']], popup='{0} {1}'.format(x,row['Code']),\n icon=folium.Icon(color=y)).add_to(stn_map)\n \n ",
"_____no_output_____"
],
[
"stn_map\nstn_map.save('stn_map.html')",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
d0b818cc160a6017ddeb8238f542a5b2191964b9 | 237,983 | ipynb | Jupyter Notebook | DEPRECATED/yemen_time_series_data_analysis.ipynb | Haleem85/Yemen | 2d09c35ff4887d70542b00b1655ab699fcabe897 | [
"MIT"
] | 1 | 2019-02-23T14:34:42.000Z | 2019-02-23T14:34:42.000Z | DEPRECATED/yemen_time_series_data_analysis.ipynb | Haleem85/Yemen | 2d09c35ff4887d70542b00b1655ab699fcabe897 | [
"MIT"
] | null | null | null | DEPRECATED/yemen_time_series_data_analysis.ipynb | Haleem85/Yemen | 2d09c35ff4887d70542b00b1655ab699fcabe897 | [
"MIT"
] | 3 | 2019-02-23T14:40:49.000Z | 2020-10-08T12:26:13.000Z | 39.336033 | 548 | 0.3302 | [
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.preprocessing import StandardScaler\nfrom scipy.stats import kurtosis\nfrom sklearn.decomposition import PCA\nimport seaborn as sns\nfrom scipy.stats import pearsonr\n%matplotlib",
"Using matplotlib backend: Qt5Agg\n"
],
[
"gov_pop_area_data = pd.read_excel('/Users/Rohil/Documents/iGEM/yemen/gov_area_pop_data.xlsx')",
"_____no_output_____"
],
[
"gov_pop_area_data = gov_pop_area_data[gov_pop_area_data.iso != 'YE-HD']",
"_____no_output_____"
],
[
"gov_pop_area_data.head()",
"_____no_output_____"
],
[
"cholera_case_crosstab = pd.read_csv(r'C:\\Users\\Rohil\\Documents\\iGEM\\yemen\\cholera_epi_data\\yemen_cholera_case_data_differenced.csv', dayfirst = True)",
"_____no_output_____"
],
[
"cholera_case_crosstab.tail()",
"_____no_output_____"
],
[
"norm_cholera_case_crosstab = cholera_case_crosstab",
"_____no_output_____"
],
[
"for index, row in gov_pop_area_data[['iso', 'population']].iterrows():\n norm_cholera_case_crosstab[row.iso] = (norm_cholera_case_crosstab[row.iso] * 10000) / row.population",
"_____no_output_____"
],
[
"norm_cholera_case_crosstab.tail()",
"_____no_output_____"
],
[
"cholera_death_crosstab = pd.read_csv(r'C:\\Users\\Rohil\\Documents\\iGEM\\yemen\\cholera_epi_data\\yemen_cholera_death_data_differenced.csv', dayfirst = True)",
"_____no_output_____"
],
[
"cholera_death_crosstab.head()",
"_____no_output_____"
],
[
"norm_cholera_death_crosstab = cholera_death_crosstab",
"_____no_output_____"
],
[
"for index, row in gov_pop_area_data[['iso', 'population']].iterrows():\n norm_cholera_death_crosstab[row.iso] = (norm_cholera_death_crosstab[row.iso] * 10000) / row.population",
"_____no_output_____"
],
[
"norm_cholera_death_crosstab.head()",
"_____no_output_____"
],
[
"mean_rainfall_crosstab = pd.read_csv(r'C:\\Users\\Rohil\\Documents\\iGEM\\yemen\\rainfall\\yemen_daily_mean_rainfall_crosstab.csv', dayfirst = True)",
"_____no_output_____"
],
[
"max_rainfall_crosstab = pd.read_csv(r'C:\\Users\\Rohil\\Documents\\iGEM\\yemen\\rainfall\\yemen_daily_max_rainfall_crosstab.csv', dayfirst = True)",
"_____no_output_____"
],
[
"mean_rainfall_crosstab.head()",
"_____no_output_____"
],
[
"max_rainfall_crosstab.head()",
"_____no_output_____"
],
[
"cases_unstacked = norm_cholera_case_crosstab.set_index('date').unstack().reset_index()\ncases_unstacked.columns = ['gov_iso', 'date', 'new_cases']",
"_____no_output_____"
],
[
"deaths_unstacked = norm_cholera_death_crosstab.set_index('date').unstack().reset_index()\ndeaths_unstacked.columns = ['gov_iso', 'date', 'new_deaths']",
"_____no_output_____"
],
[
"max_rainfall_unstacked = max_rainfall_crosstab.set_index('date').unstack().reset_index()\nmax_rainfall_unstacked.columns = ['gov_iso', 'date', 'max_rainfall']",
"_____no_output_____"
],
[
"mean_rainfall_unstacked = mean_rainfall_crosstab.set_index('date').unstack().reset_index()\nmean_rainfall_unstacked.columns = ['gov_iso', 'date', 'mean_rainfall']",
"_____no_output_____"
],
[
"cases_unstacked.shape",
"_____no_output_____"
],
[
"cases_unstacked.head()",
"_____no_output_____"
],
[
"deaths_unstacked.shape",
"_____no_output_____"
],
[
"deaths_unstacked.head()",
"_____no_output_____"
],
[
"mean_rainfall_unstacked.shape",
"_____no_output_____"
],
[
"## date formatting has been fixed\nmean_rainfall_unstacked.head()",
"_____no_output_____"
],
[
"mean_rainfall_unstacked.date.tail()",
"_____no_output_____"
],
[
"cases_unstacked.date.tail()",
"_____no_output_____"
],
[
"deaths_unstacked.date.tail()",
"_____no_output_____"
],
[
"case_death_rainfall_data = cases_unstacked.merge(deaths_unstacked, on =['date', 'gov_iso']).merge(mean_rainfall_unstacked, on =['date', 'gov_iso'], how = 'left')",
"_____no_output_____"
],
[
"case_death_rainfall_data.date = pd.to_datetime(case_death_rainfall_data.date, dayfirst = True)",
"_____no_output_____"
],
[
"case_death_rainfall_data.sort_values(by = 'date')",
"_____no_output_____"
],
[
"# YE-HD-AL refers to Al Mukulla\nneighboring_gov_dict = {\"YE-SA\" : [\"YE-SN\"], \n \"YE-AB\" : [\"YE-LA\", \"YE-SH\", \"YE-BA\"], \n \"YE-AD\" : [\"YE-LA\"], \n \"YE-DA\" : [\"YE-LA\", \"YE-TA\", \"YE-IB\", \"YE-BA\"], \n \"YE-BA\" : [\"YE-DH\", \"YE-IB\", \"YE-DA\", \"YE-AB\", \"YE-SH\", \"YE-MA\", \"YE-SN\"], \n \"YE-HU\" : [\"YE-HJ\", \"YE-MW\", \"YE-SN\", \"YE-RA\", \"YE-DH\", \"YE-TA\"], \n \"YE-JA\" : [\"YE-MA\", \"YE-SN\", \"YE-AM\", \"YE-SD\"], \n \"YE-MR\" : [\"YE-HD-AL\"], \n \"YE-MW\" : [\"YE-HU\", \"YE-HJ\", \"YE-AM\", \"YE-SN\"], \n \"YE-AM\" : [\"YE-HJ\", \"YE-SD\", \"YE-JA\", \"YE-SN\", \"YE-MW\"], \n \"YE-DH\" : [\"YE-IB\", \"YE-RA\", \"YE-SN\", \"YE-BA\"], \n \"YE-HD-AL\" : [\"YE-SH\", \"YE-MR\"], \n \"YE-HJ\" : [\"YE-MW\", \"YE-HU\", \"YE-MR\"], \n \"YE-IB\" : [\"YE-TA\", \"YE-HU\", \"YE-DH\", \"YE-BA\", \"YE-DA\"], \n \"YE-LA\" : [\"YE-AD\", \"YE-TA\", \"YE-DA\", \"YE-BA\", \"YE-AB\"], \n \"YE-MA\" : [\"YE-BA\", \"YE-SN\", \"YE-JA\", \"YE-SH\"], \n \"YE-RA\" : [\"YE-DH\", \"YE-HU\", \"YE-SN\"], \n \"YE-SD\" : [\"YE-HJ\", \"YE-AM\", \"YE-JA\"], \n \"YE-SN\" : [\"YE-BA\", \"YE-DH\", \"YE-RA\", \"YE-MW\", \"YE-AM\", \"YE-JA\", \"YE-MA\"], \n \"YE-SH\" : [\"YE-AB\", \"YE-BA\", \"YE-MA\", \"YE-HD-AL\"], \n \"YE-TA\" : [\"YE-LA\", \"YE-DA\", \"YE-IB\", \"YE-HU\"]}",
"_____no_output_____"
],
[
"def get_past_days_features(row, var, daysback):\n stock_data = full_data[full_data.stock_id == row.stock_id].set_index('date')\n x_days_date = row.date - pd.to_timedelta(daysback, unit='d')\n relevant_stock_data = stock_data.loc[(stock_data.index >= x_days_date) & (stock_data.index < row.date)].sort_index()\n return (pd.Series([np.mean(relevant_stock_data[var]), np.max(relevant_stock_data[var]), kurtosis(relevant_stock_data[var])]))",
"_____no_output_____"
],
[
"def get_past_days_features(row, var, daysback):\n other_stock_data = full_data[full_data.stock_id.isin(neighboring_stocks[row.stock_id])].set_index('date')\n x_days_date = row.date - pd.to_timedelta(daysback, unit='d')\n relevant_other_stock_data = other_stock_data.loc[(other_stock_data.index >= x_days_date) & (other_stock_data.index < row.date)].sort_index()\n return (pd.Series([np.mean(relevant_other_stock_data[var]), np.max(relevant_other_stock_data[var]), kurtosis(relevant_other_stock_data[var])]))",
"_____no_output_____"
],
[
"def get_past_days_features(row, var, daysback):\n if 'rainfall' in var:\n rainfall_df = mean_rainfall_unstacked\n rainfall_df.date = pd.to_datetime(rainfall_df.date, dayfirst = True)\n gov_data = rainfall_df[rainfall_df.gov_iso == row.gov_iso].set_index('date')\n x_days_date = row.date - pd.to_timedelta(daysback, unit='d')\n relevant_gov_data = gov_data.loc[(gov_data.index >= x_days_date) & (gov_data.index < row.date)].sort_index()\n return (pd.Series([np.mean(relevant_gov_data[var]), np.max(relevant_gov_data[var]), kurtosis(relevant_gov_data[var])])) \n else: \n gov_data = case_death_rainfall_data[case_death_rainfall_data.gov_iso == row.gov_iso].set_index('date')\n x_days_date = row.date - pd.to_timedelta(daysback, unit='d')\n relevant_gov_data = gov_data.loc[(gov_data.index >= x_days_date) & (gov_data.index < row.date)].sort_index()\n return (pd.Series([np.mean(relevant_gov_data[var]), np.max(relevant_gov_data[var]), kurtosis(relevant_gov_data[var])]))",
"_____no_output_____"
],
[
"def get_neighbor_past_days_features(row, var, daysback):\n if 'rainfall' in var:\n rainfall_df = mean_rainfall_unstacked\n rainfall_df.date = pd.to_datetime(rainfall_df.date, dayfirst = True)\n other_gov_data = rainfall_df[rainfall_df.gov_iso.isin(neighboring_gov_dict[row.gov_iso])].set_index('date')\n x_days_date = row.date - pd.to_timedelta(daysback, unit='d')\n relevant_other_gov_data = other_gov_data.loc[(other_gov_data.index >= x_days_date) & (other_gov_data.index < row.date)].sort_index()\n return (pd.Series([np.mean(relevant_other_gov_data[var]), np.max(relevant_other_gov_data[var]), kurtosis(relevant_other_gov_data[var])])) \n else: \n other_gov_data = case_death_rainfall_data[case_death_rainfall_data.gov_iso.isin(neighboring_gov_dict[row.gov_iso])].set_index('date')\n x_days_date = row.date - pd.to_timedelta(daysback, unit='d')\n relevant_other_gov_data = other_gov_data.loc[(other_gov_data.index >= x_days_date) & (other_gov_data.index < row.date)].sort_index()\n return (pd.Series([np.mean(relevant_other_gov_data[var]), np.max(relevant_other_gov_data[var]), kurtosis(relevant_other_gov_data[var])]))",
"_____no_output_____"
],
[
"past_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 7), axis = 1)\npast_week_cases.columns = ['mean_past_week_cases', 'max_past_week_cases', 'kurtosis_past_week_cases']\nneighbor_past_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 7), axis = 1)\nneighbor_past_week_cases.columns = ['neighbor_mean_past_week_cases', 'neighbor_max_past_week_cases', 'neighbor_kurtosis_past_week_cases']\n\npast_2_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 14), axis = 1)\npast_2_week_cases.columns = ['mean_past_2_week_cases', 'max_past_2_week_cases', 'kurtosis_past_2_week_cases']\nneighbor_past_2_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 14), axis = 1)\nneighbor_past_2_week_cases.columns = ['neighbor_mean_past_2_week_cases', 'neighbor_max_past_2_week_cases', 'neighbor_kurtosis_past_2_week_cases']\n\npast_3_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 21), axis = 1)\npast_3_week_cases.columns = ['mean_past_3_week_cases', 'max_past_3_week_cases', 'kurtosis_past_3_week_cases']\nneighbor_past_3_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 21), axis = 1)\nneighbor_past_3_week_cases.columns = ['neighbor_mean_past_3_week_cases', 'neighbor_max_past_3_week_cases', 'neighbor_kurtosis_past_3_week_cases']\n\npast_month_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 30), axis = 1)\npast_month_cases.columns = ['mean_past_month_cases', 'max_past_month_cases', 'kurtosis_past_month_cases']\nneighbor_past_month_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 30), axis = 1)\nneighbor_past_month_cases.columns = ['neighbor_mean_past_month_cases', 'neighbor_max_past_month_cases', 'neighbor_kurtosis_past_month_cases']\n\npast_6_week_cases = case_death_rainfall_data.apply(get_past_days_features, args = ('new_cases', 42), axis = 1)\npast_6_week_cases.columns = ['mean_past_6_week_cases', 'max_past_6_week_cases', 'kurtosis_past_6_week_cases']\nneighbor_past_6_week_cases = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_cases', 42), axis = 1)\nneighbor_past_6_week_cases.columns = ['neighbor_mean_past_6_week_cases', 'neighbor_max_past_6_week_cases', 'neighbor_kurtosis_past_6_week_cases']",
"_____no_output_____"
],
[
"past_week_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 7), axis = 1)\npast_week_deaths.columns = ['mean_past_week_deaths', 'max_past_week_deaths', 'kurtosis_past_week_deaths']\nneighbor_past_week_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 7), axis = 1)\nneighbor_past_week_deaths.columns = ['neighbor_mean_past_week_deaths', 'neighbor_max_past_week_deaths', 'neighbor_kurtosis_past_week_deaths']\n\npast_2_week_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 14), axis = 1)\npast_2_week_deaths.columns = ['mean_past_2_week_deaths', 'max_past_2_week_deaths', 'kurtosis_past_2_week_deaths']\nneighbor_past_2_week_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 14), axis = 1)\nneighbor_past_2_week_deaths.columns = ['neighbor_mean_past_2_week_deaths', 'neighbor_max_past_2_week_deaths', 'neighbor_kurtosis_past_2_week_deaths']\n\npast_month_deaths = case_death_rainfall_data.apply(get_past_days_features, args = ('new_deaths', 30), axis = 1)\npast_month_deaths.columns = ['mean_past_month_deaths', 'max_past_month_deaths', 'kurtosis_past_month_deaths']\nneighbor_past_month_deaths = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('new_deaths', 30), axis = 1)\nneighbor_past_month_deaths.columns = ['neighbor_mean_past_month_deaths', 'neighbor_max_past_month_deaths', 'neighbor_kurtosis_past_month_deaths']",
"_____no_output_____"
],
[
"past_week_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 7), axis = 1)\npast_week_rainfall.columns = ['mean_past_week_rainfall', 'max_past_week_rainfall', 'kurtosis_past_week_rainfall']\nneighbor_past_week_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 7), axis = 1)\nneighbor_past_week_rainfall.columns = ['neighbor_mean_past_week_rainfall', 'neighbor_max_past_week_rainfall', 'neighbor_kurtosis_past_week_rainfall']\n\npast_2_week_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 14), axis = 1)\npast_2_week_rainfall.columns = ['mean_past_2_week_rainfall', 'max_past_2_week_rainfall', 'kurtosis_past_2_week_rainfall']\nneighbor_past_2_week_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 14), axis = 1)\nneighbor_past_2_week_rainfall.columns = ['neighbor_mean_past_2_week_rainfall', 'neighbor_max_past_2_week_rainfall', 'neighbor_kurtosis_past_2_week_rainfall']\n\npast_month_rainfall = case_death_rainfall_data.apply(get_past_days_features, args = ('mean_rainfall', 30), axis = 1)\npast_month_rainfall.columns = ['mean_past_month_rainfall', 'max_past_month_rainfall', 'kurtosis_past_month_rainfall']\nneighbor_past_month_rainfall = case_death_rainfall_data.apply(get_neighbor_past_days_features, args = ('mean_rainfall', 30), axis = 1)\nneighbor_past_month_rainfall.columns = ['neighbor_mean_past_month_rainfall', 'neighbor_max_past_month_rainfall', 'neighbor_kurtosis_past_month_rainfall']",
"_____no_output_____"
],
[
"training_data = pd.concat([case_death_rainfall_data[['gov_iso', 'date', 'weekly_cases']], \n past_week_cases, past_2_week_cases, past_month_cases, neighbor_past_week_cases, neighbor_past_2_week_cases, neighbor_past_month_cases,\n past_week_deaths, past_2_week_deaths, past_month_deaths, neighbor_past_week_deaths, neighbor_past_2_week_deaths, neighbor_past_month_deaths,\n past_week_rainfall, past_2_week_rainfall, past_month_rainfall, neighbor_past_week_rainfall, neighbor_past_2_week_rainfall, neighbor_past_month_rainfall], axis = 1)",
"_____no_output_____"
],
[
"training_data.to_csv('/Users/Rohil/Documents/iGEM/yemen/full_feature_data.csv', index = False)",
"_____no_output_____"
],
[
"col_list = []\nfor col in training_data.columns:\n if ('max' not in col) and ('kurtosis' not in col) & ('deaths' not in col):\n col_list.append(col)",
"_____no_output_____"
],
[
"# want to have at least 7 days of data for most of these examples\ntrunc_training_data = training_data[col_list]\ntrunc_training_data = trunc_training_data[(trunc_training_data['date'] > '2017-05-30')].sort_values('date')",
"_____no_output_____"
],
[
"features = trunc_training_data.iloc[:,3:].columns.tolist()\ntarget = trunc_training_data.iloc[:,2].name",
"_____no_output_____"
],
[
"correlations = {}\nfor f in features:\n data_temp = trunc_training_data[[f,target]]\n x1 = data_temp[f].values\n x2 = data_temp[target].values\n key = f + ' vs ' + target\n correlations[key] = pearsonr(x1,x2)[0]",
"_____no_output_____"
],
[
"data_correlations = pd.DataFrame(correlations, index=['Value']).T\ndata_correlations.loc[data_correlations['Value'].abs().sort_values(ascending=False).index]",
"_____no_output_____"
],
[
"trunc_training_data = pd.concat([trunc_training_data, pd.get_dummies(trunc_training_data.gov_iso).sort_index()], axis=1)",
"_____no_output_____"
],
[
"trunc_training_data.to_csv('/Users/Rohil/Documents/iGEM/yemen/prelim_training_data.csv', index = False)",
"_____no_output_____"
],
[
"trunc_training_data[trunc_training_data.isnull().any(axis=1)]",
"_____no_output_____"
],
[
"trunc_training_data.shape",
"_____no_output_____"
],
[
"trunc_training_data.head()",
"_____no_output_____"
],
[
"whole_standard_scaler = StandardScaler()",
"_____no_output_____"
],
[
"trunc_training_features = trunc_training_data.iloc[:,3:]",
"_____no_output_____"
],
[
"trunc_training_features.shape",
"_____no_output_____"
],
[
"norm_features = whole_standard_scaler.fit_transform(trunc_training_features)",
"_____no_output_____"
],
[
"pca = PCA(n_components = 33)",
"_____no_output_____"
],
[
"pca.fit(norm_features)",
"_____no_output_____"
],
[
"pca.explained_variance_ratio_",
"_____no_output_____"
],
[
"pd.DataFrame(pca.components_, columns = trunc_training_features.columns)",
"_____no_output_____"
],
[
"pca.components_",
"_____no_output_____"
],
[
"sns.heatmap(np.log(pca.inverse_transform(np.eye(12))))",
"C:\\Users\\Rohil\\Anaconda3\\lib\\site-packages\\ipykernel_launcher.py:1: RuntimeWarning: invalid value encountered in log\n \"\"\"Entry point for launching an IPython kernel.\n"
],
[
"# plots of normalized cases x days back vs today\nfor column in trunc_training_features.columns:\n fig, ax = plt.subplots(1,1)\n ax.scatter(trunc_training_features[column], trunc_training_data['weekly_cases'])\n ax.set_ylabel('weekly cholera cases')\n ax.set_xlabel(column)\n fig.savefig('/Users/Rohil/Documents/iGEM/yemen/feature_engineering/old/' + column + '_vs_cases.png')\n plt.close()",
"_____no_output_____"
],
[
"norm_features = pd.DataFrame(data=norm_features, columns = trunc_training_features.columns)",
"_____no_output_____"
],
[
"norm_features",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
d0b851ff1884b27498586b9302b1637e0c1b777b | 11,853 | ipynb | Jupyter Notebook | solution-image-blending.ipynb | tek5030/lab-image-blending-py | 30ac2c8bd4d57759188e3f419aab1ed489d2dff9 | [
"BSD-3-Clause"
] | null | null | null | solution-image-blending.ipynb | tek5030/lab-image-blending-py | 30ac2c8bd4d57759188e3f419aab1ed489d2dff9 | [
"BSD-3-Clause"
] | null | null | null | solution-image-blending.ipynb | tek5030/lab-image-blending-py | 30ac2c8bd4d57759188e3f419aab1ed489d2dff9 | [
"BSD-3-Clause"
] | null | null | null | 31.028796 | 197 | 0.577828 | [
[
[
"# Implement image blending\n\nWe will start by importing libraries and defining a couple of functions for displaying images using matplotlib.",
"_____no_output_____"
]
],
[
[
"import cv2\nimport matplotlib.pyplot as plt\nimport numpy as np\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = [20, 10]\n\ndef showResult(title, img):\n # Colour images in OpenCV are given in BGR, but matplotlib expects RGB.\n # We therefore need to convert the OpenCV images.\n plt.imshow(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))\n plt.title(title)\n plt.show()\n\ndef showResultsSideBySide(title1, img1, title2, img2):\n # Display the original and the transformed image\n axes = plt.subplots(1, 2)[1]\n ax1, ax2 = axes\n ax1.set_title(title1)\n ax2.set_title(title2)\n ax1.imshow(cv2.cvtColor(img1, cv2.COLOR_BGR2RGB))\n ax2.imshow(cv2.cvtColor(img2, cv2.COLOR_BGR2RGB))\n plt.show()",
"_____no_output_____"
]
],
[
[
"## 1. Load and convert the source images\nNow let's do some image processing!\n\nFirst, we need to read the two images we want to blend.\n\n <table cellspacing=\"0\" cellpadding=\"0\"><tr>\n <td> <img src=\"img/thumb_lion.png\" alt=\"Lion\" style=\"width: 200px;\"/> </td>\n <td> <img src=\"img/thumb_tiger.png\" alt=\"Tiger\" style=\"width: 200px;\"/> </td>\n <td> <img src=\"img/thumb_white_tiger.png\" alt=\"White tiger\" style=\"width: 200px;\"/> </td>\n </tr></table>\n\n- Read two images (choose two of the images given in the `data` directory)\n - Hint: [cv::imread(...)](https://docs.opencv.org/4.0.1/d4/da8/group__imgcodecs.html#ga288b8b3da0892bd651fce07b3bbd3a56)\n\n- Convert the images to `float32` and scale the pixel values so that they will lie in the interval [0, 1]\n - Hint: [numpy.float32](https://numpy.org/doc/stable/reference/arrays.scalars.html?#numpy.float32)",
"_____no_output_____"
]
],
[
[
"# Load the images.\nimg_01_fname = \"data/tiger.png\"\nimg_01 = cv2.imread(img_01_fname)\nimg_01 = np.float32(img_01) * (1.0/255.0)\n\nimg_02_fname = \"data/white_tiger.png\"\nimg_02 = cv2.imread(img_02_fname)\nimg_02 = np.float32(img_02) * (1.0/255.0)\n\n# Show the loaded images.\nshowResultsSideBySide(img_01_fname, img_01, img_02_fname, img_02)",
"_____no_output_____"
]
],
[
[
"## 2. Create an image with weights for blending\nNow we need to define how the two images should be blended together.\nWe will do this by constructing a weight image with weights $w(u, v) \\in [0, 1].$\n\nA weight of 1 means that the blended pixel will be equal to the corresponding pixel in image 1, while a weight of 0.5 means that the resulting pixel is an equally large mix of both images.\n\n### a) Create the weight image:\n - The size is equal to the size of the input images\n - It should have 3 channels given in 32-bit floating point\n - The left half of the image should be black (pixel value 0.0 in all channels)\n - The right half of the image should be white (pixel value 1.0 in all channels)\n\n### b) Make a ramp (a smooth gradient) in the transition between black and white\n - Hint: [cv::blur](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#ga8c45db9afe636703801b0b2e440fce37)\n\n ",
"_____no_output_____"
]
],
[
[
"# Construct a half black, half white image.\nweights = np.zeros(img_01.shape, dtype=np.float32)\nhalf_image_width = int(0.5 * weights.shape[1])\nweights[:, :half_image_width] = (1., 1., 1.)\n\n# Create a ramp between the two halves.\nramp_width = 50\nweights = cv2.blur(weights, (ramp_width+1, ramp_width+1))\n\n# Visualise the weights.\nshowResult(\"weights\", weights)",
"_____no_output_____"
]
],
[
[
"## 3. Simple linear blending\nThe next step is to implement functionality for simple linear blending, where the two images are mixed according to the weight image.\n\n### a) Implement linear blending of two images using the weights\n- $res = w \\cdot img_1 + (1-w) \\cdot img_2$\n- Tip: You can solve this step using only image operations, without writing any loops.\n\n### b) Run the code and check that the result looks reasonable\n- Try changing the ramp size `ramp_width`. What happens?\n- Try making the blend as smooth and visually pleasing as possible.",
"_____no_output_____"
]
],
[
[
"def linearBlending(img1, img2, mask):\n return img1 * mask + img2 * (1.-mask)\n\n# Test linear blending.\nlinear_blend = linearBlending(img_01, img_02, weights)\nshowResult('linear_blend', linear_blend)",
"_____no_output_____"
]
],
[
[
"## 4. Laplace blending\nTo demonstrate the difference between simple linear blending, and scale-aware blending, we will now implement and test Laplace blending.\nWe will even get to play around with scale pyramids!\n\nRecall from the lecture that Laplace blending performs linear blending at different stages in the laplacian pyramid for an image (at different scales):\n\n\n\nFirst we convert the image to a laplacian pyramid, then we perform the linear blending, and finally we reconstruct the blended laplacian into the resulting blended image.\n\n### a) Construct a Gaussian pyramid\n\n\n\n- Hint: Use [cv::pyrDown()](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#gaf9bba239dfca11654cb7f50f889fc2ff)\n",
"_____no_output_____"
]
],
[
[
"def constructGaussianPyramid(img):\n # Construct the pyramid starting with the original image.\n pyr = [img]\n\n # Add new downscaled images to the pyramid\n # until image width is <= 16 pixels\n while pyr[-1].shape[1] > 16:\n pyr.append(cv2.pyrDown(pyr[-1]))\n\n return pyr\n",
"_____no_output_____"
]
],
[
[
"### b) Construct a Laplacian pyramid\n\n\n\n- Hint: Use [cv::pyrUp()](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#gada75b59bdaaca411ed6fee10085eb784)\n",
"_____no_output_____"
]
],
[
[
"def constructLaplacianPyramid(img):\n pyr = constructGaussianPyramid(img)\n for i in range(len(pyr)-1):\n pyr[i] -= cv2.pyrUp(pyr[i+1], dstsize=pyr[i].shape[0:2])\n\n return pyr",
"_____no_output_____"
]
],
[
[
"### c) Implement function for collapsing the Laplacian pyramid\n\n\n- Hint: Use [cv::pyrUp()](https://docs.opencv.org/4.5.5/d4/d86/group__imgproc__filter.html#gada75b59bdaaca411ed6fee10085eb784)\n",
"_____no_output_____"
]
],
[
[
"def collapsePyramid(pyr):\n for i in range(len(pyr)-2, -1, -1):\n pyr[i] += cv2.pyrUp(pyr[i+1], dstsize=pyr[i].shape[0:2])\n \n return pyr[0]",
"_____no_output_____"
]
],
[
[
"### d) Implement Laplacian blending\n- Construct a Gaussian pyramid for the weights.\n- Construct Laplacian pyramids for the images.\n- Blend the images using `linearBlending()` on each pyramid level.\n- Reconstruct the blended image by collapsing the blended pyramid.",
"_____no_output_____"
]
],
[
[
"def laplaceBlending(img1, img2, mask):\n # Construct a gaussian pyramid of the mask image.\n pyr_mask = constructGaussianPyramid(mask)\n\n # Construct a laplacian pyramid of each of the images.\n pyr_img1 = constructLaplacianPyramid(img1)\n pyr_img2 = constructLaplacianPyramid(img2)\n\n # Blend the laplacian pyramids according to the corresponding weight pyramid.\n pyr_blend = []\n for img1_lvl, img2_lvl, mask_lvl in zip(pyr_img1, pyr_img2, pyr_mask):\n pyr_blend.append(\n linearBlending(img1_lvl, img2_lvl, mask_lvl)\n )\n\n # Collapse the blended Laplacian pyramid.\n return collapsePyramid(pyr_blend)",
"_____no_output_____"
]
],
[
[
"### e) Check that the results look reasonable\n- Test Laplace blending\n- Compare the results with linear blending.\n- What happens when you reduce the ramp size down to a very steep gradient?",
"_____no_output_____"
]
],
[
[
"lap_blend = laplaceBlending(img_01, img_02, weights)\nshowResult('blend', lap_blend)",
"_____no_output_____"
]
],
[
[
"### f) Experiments\nTry other images\n - Capture images using the camera\n - Download images from the internet\n\nTry other weight masks\n - Circles\n - Other shapes",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
d0b85c4509ee67b4c7efcaa050147e494970b4a7 | 15,886 | ipynb | Jupyter Notebook | M6_Python_for_Data_Science_AI_Development/Week4/files/PY0101EN-5-2-Numpy2D.ipynb | cmaroblesg/IBM_DevOps-_and_Software_Engineering | b943be2e5774e2f54e5814ff9c7bd11c7a178d47 | [
"MIT"
] | null | null | null | M6_Python_for_Data_Science_AI_Development/Week4/files/PY0101EN-5-2-Numpy2D.ipynb | cmaroblesg/IBM_DevOps-_and_Software_Engineering | b943be2e5774e2f54e5814ff9c7bd11c7a178d47 | [
"MIT"
] | null | null | null | M6_Python_for_Data_Science_AI_Development/Week4/files/PY0101EN-5-2-Numpy2D.ipynb | cmaroblesg/IBM_DevOps-_and_Software_Engineering | b943be2e5774e2f54e5814ff9c7bd11c7a178d47 | [
"MIT"
] | null | null | null | 15,886 | 15,886 | 0.659323 | [
[
[
"<center>\n <img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n\n# 2D Numpy in Python\n\nEstimated time needed: **20** minutes\n\n## Objectives\n\nAfter completing this lab you will be able to:\n\n* Operate comfortably with `numpy`\n* Perform complex operations with `numpy`\n",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-thttps://op/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01: 20px\">\n <ul>\n <li><a href=\"https://create/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Create a 2D Numpy Array</a></li>\n <li><a href=\"https://access/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Accessing different elements of a Numpy Array</a></li>\n <li><a href=\"op\">Basic Operations</a></li>\n </ul>\n\n</div>\n\n<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"create\">Create a 2D Numpy Array</h2>\n",
"_____no_output_____"
]
],
[
[
"# Import the libraries\n\nimport numpy as np \nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Consider the list <code>a</code>, which contains three nested lists **each of equal size**.\n",
"_____no_output_____"
]
],
[
[
"# Create a list\n\na = [[11, 12, 13], [21, 22, 23], [31, 32, 33]]\na",
"_____no_output_____"
]
],
[
[
"We can cast the list to a Numpy Array as follows:\n",
"_____no_output_____"
]
],
[
[
"# Convert list to Numpy Array\n# Every element is the same type\n\nA = np.array(a)\nA",
"_____no_output_____"
]
],
[
[
"We can use the attribute <code>ndim</code> to obtain the number of axes or dimensions, referred to as the rank.\n",
"_____no_output_____"
]
],
[
[
"# Show the numpy array dimensions\n\nA.ndim",
"_____no_output_____"
]
],
[
[
"Attribute <code>shape</code> returns a tuple corresponding to the size or number of each dimension.\n",
"_____no_output_____"
]
],
[
[
"# Show the numpy array shape\n\nA.shape",
"_____no_output_____"
]
],
[
[
"The total number of elements in the array is given by the attribute <code>size</code>.\n",
"_____no_output_____"
]
],
[
[
"# Show the numpy array size\n\nA.size",
"_____no_output_____"
]
],
[
[
"<hr>\n",
"_____no_output_____"
],
[
"<h2 id=\"access\">Accessing different elements of a Numpy Array</h2>\n",
"_____no_output_____"
],
[
"We can use rectangular brackets to access the different elements of the array. The correspondence between the rectangular brackets and the list and the rectangular representation is shown in the following figure for a 3x3 array:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoEg.png\" width=\"500\" />\n",
"_____no_output_____"
],
[
"We can access the 2nd-row, 3rd column as shown in the following figure:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoFT.png\" width=\"400\" />\n",
"_____no_output_____"
],
[
"We simply use the square brackets and the indices corresponding to the element we would like:\n",
"_____no_output_____"
]
],
[
[
"# Access the element on the second row and third column\n\nA[1, 2]",
"_____no_output_____"
]
],
[
[
"We can also use the following notation to obtain the elements:\n",
"_____no_output_____"
]
],
[
[
"# Access the element on the second row and third column\n\nA[1][2]",
"_____no_output_____"
]
],
[
[
"Consider the elements shown in the following figure\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoFF.png\" width=\"400\" />\n",
"_____no_output_____"
],
[
"We can access the element as follows:\n",
"_____no_output_____"
]
],
[
[
"# Access the element on the first row and first column\n\nA[0][0]",
"_____no_output_____"
]
],
[
[
"We can also use slicing in numpy arrays. Consider the following figure. We would like to obtain the first two columns in the first row\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoFSF.png\" width=\"400\" />\n",
"_____no_output_____"
],
[
"This can be done with the following syntax:\n",
"_____no_output_____"
]
],
[
[
"# Access the element on the first row and first and second columns\n\nA[0][0:2]",
"_____no_output_____"
]
],
[
[
"Similarly, we can obtain the first two rows of the 3rd column as follows:\n",
"_____no_output_____"
]
],
[
[
"# Access the element on the first and second rows and third column\n\nA[0:2, 2]",
"_____no_output_____"
]
],
[
[
"Corresponding to the following figure:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/2D_numpy.png\" width=\"550\"><br />\n",
"_____no_output_____"
],
[
"<h2 id=\"op\">Basic Operations</h2>\n",
"_____no_output_____"
],
[
"We can also add arrays. The process is identical to matrix addition. Matrix addition of <code>X</code> and <code>Y</code> is shown in the following figure:\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoAdd.png\" width=\"500\" />\n",
"_____no_output_____"
],
[
"The numpy array is given by <code>X</code> and <code>Y</code>\n",
"_____no_output_____"
]
],
[
[
"# Create a numpy array X\n\nX = np.array([[1, 0], [0, 1]]) \nX",
"_____no_output_____"
],
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
]
],
[
[
"We can add the numpy arrays as follows.\n",
"_____no_output_____"
]
],
[
[
"# Add X and Y\n\nZ = X + Y\nZ",
"_____no_output_____"
]
],
[
[
"Multiplying a numpy array by a scaler is identical to multiplying a matrix by a scaler. If we multiply the matrix <code>Y</code> by the scaler 2, we simply multiply every element in the matrix by 2, as shown in the figure.\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoDb.png\" width=\"500\" />\n",
"_____no_output_____"
],
[
"We can perform the same operation in numpy as follows\n",
"_____no_output_____"
]
],
[
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
],
[
"# Multiply Y with 2\n\nZ = 2 * Y\nZ",
"_____no_output_____"
]
],
[
[
"Multiplication of two arrays corresponds to an element-wise product or <em>Hadamard product</em>. Consider matrix <code>X</code> and <code>Y</code>. The Hadamard product corresponds to multiplying each of the elements in the same position, i.e. multiplying elements contained in the same color boxes together. The result is a new matrix that is the same size as matrix <code>Y</code> or <code>X</code>, as shown in the following figure.\n",
"_____no_output_____"
],
[
"<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%205/images/NumTwoMul.png\" width=\"500\" />\n",
"_____no_output_____"
],
[
"We can perform element-wise product of the array <code>X</code> and <code>Y</code> as follows:\n",
"_____no_output_____"
]
],
[
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
],
[
"# Create a numpy array X\n\nX = np.array([[1, 0], [0, 1]]) \nX",
"_____no_output_____"
],
[
"# Multiply X with Y\n\nZ = X * Y\nZ",
"_____no_output_____"
]
],
[
[
"We can also perform matrix multiplication with the numpy arrays <code>A</code> and <code>B</code> as follows:\n",
"_____no_output_____"
],
[
"First, we define matrix <code>A</code> and <code>B</code>:\n",
"_____no_output_____"
]
],
[
[
"# Create a matrix A\n\nA = np.array([[0, 1, 1], [1, 0, 1]])\nA",
"_____no_output_____"
],
[
"# Create a matrix B\n\nB = np.array([[1, 1], [1, 1], [-1, 1]])\nB",
"_____no_output_____"
]
],
[
[
"We use the numpy function <code>dot</code> to multiply the arrays together.\n",
"_____no_output_____"
]
],
[
[
"# Calculate the dot product\n\nZ = np.dot(A,B)\nZ",
"_____no_output_____"
],
[
"# Calculate the sine of Z\n\nnp.sin(Z)",
"_____no_output_____"
]
],
[
[
"We use the numpy attribute <code>T</code> to calculate the transposed matrix\n",
"_____no_output_____"
]
],
[
[
"# Create a matrix C\n\nC = np.array([[1,1],[2,2],[3,3]])\nC",
"_____no_output_____"
],
[
"# Get the transposed of C\n\nC.T",
"_____no_output_____"
]
],
[
[
"<h2>Quiz on 2D Numpy Array</h2>\n",
"_____no_output_____"
],
[
"Consider the following list <code>a</code>, convert it to Numpy Array.\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\na = [[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]]",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA = np.array(a)\nA\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Calculate the numpy array size.\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA.size\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Access the element on the first row and first and second columns.\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nA[0][0:2]\n```\n\n</details>\n",
"_____no_output_____"
],
[
"Perform matrix multiplication with the numpy arrays <code>A</code> and <code>B</code>.\n",
"_____no_output_____"
]
],
[
[
"# Write your code below and press Shift+Enter to execute\n\nB = np.array([[0, 1], [1, 0], [1, 1], [-1, 0]])",
"_____no_output_____"
]
],
[
[
"<details><summary>Click here for the solution</summary>\n\n```python\nX = np.dot(A,B)\nX\n```\n\n</details>\n",
"_____no_output_____"
],
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. \n<hr>\n",
"_____no_output_____"
],
[
"## Author\n\n<a href=\"https://www.linkedin.com/in/joseph-s-50398b136/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\" target=\"_blank\">Joseph Santarcangelo</a>\n\n## Other contributors\n\n<a href=\"https://www.linkedin.com/in/jiahui-mavis-zhou-a4537814a?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDeveloperSkillsNetworkPY0101ENSkillsNetwork19487395-2021-01-01\">Mavis Zhou</a>\n\n## Change Log\n\n| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | ---------- | ----------------------------------------------------------- |\n| 2022-01-10 | 2.1 | Malika | Removed the readme for GitShare |\n| 2021-01-05 | 2.2 | Malika | Updated the solution for dot multiplication |\n| 2020-09-09 | 2.1 | Malika | Updated the screenshot for first two rows of the 3rd column |\n| 2020-08-26 | 2.0 | Lavanya | Moved lab to course repo in GitLab |\n| | | | |\n| | | | |\n\n<hr/>\n\n## <h3 align=\"center\"> © IBM Corporation 2020. All rights reserved. <h3/>\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.